Pre-neoplastic pancreas cells enter a partially mesenchymal state following transient TGF-β exposure
- PMID: 29713060
- PMCID: PMC6076343
- DOI: 10.1038/s41388-018-0264-6
Pre-neoplastic pancreas cells enter a partially mesenchymal state following transient TGF-β exposure
Abstract
Pancreatic ductal adenocarcinoma (PDAC) is a deadly disease and a major health problem in the United States. While the cytokine TGF-β has been implicated in PDAC development, it can exert both pro-tumorigenic and anti-tumorigenic effects that are highly context dependent and incompletely understood. Using three-dimensional (3D) cultures of KrasG12D-expressing mouse pancreatic epithelial cells we demonstrated that while exposure to exogenous TGF-β induced growth arrest of the KrasG12D cells, its subsequent removal allowed the cells to enter a hyper-proliferative, partially mesenchymal (PM), and progenitor-like state. This state was highly stable and was maintained by autocrine TGF-β signaling. While untreated KrasG12D cells formed cystic lesions in vivo, PM cells formed ductal structures resembling human PanINs, suggesting that they had attained increased oncogenic potential. Supporting this hypothesis, we determined that the PM cells share salient molecular and phenotypic features with the quasi-mesenchymal/squamous subtype of human PDAC, which has the worst prognosis of any of the recently identified subtypes. Transient pulses of TGF-β have been observed during pancreatitis, a major risk factor for PDAC. Our data suggest that transient TGF-β exposure is sufficient to induce the acquisition of stable PDAC-associated phenotypes in pre-neoplastic KrasG12D cells, providing novel molecular insight into the complex role of TGF-β in tumorigenesis.
Conflict of interest statement
The authors have no conflicts of interest to disclose.
Figures

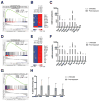
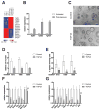

References
-
- American Cancer Society. Cancer Facts & Figures 2017. Atlanta: American Cancer Society; 2017.
-
- Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371:1039–49. - PubMed
-
- Friess H, Yamanaka Y, Buchler M, Ebert M, Beger HG, Gold LI, et al. Enhanced expression of transforming growth factor beta isoforms in pancreatic cancer correlates with decreased survival. Gastroenterology. 1993;105:1846–56. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

