Retrotransposon targeting to RNA polymerase III-transcribed genes
- PMID: 29713390
- PMCID: PMC5911963
- DOI: 10.1186/s13100-018-0119-2
Retrotransposon targeting to RNA polymerase III-transcribed genes
Abstract
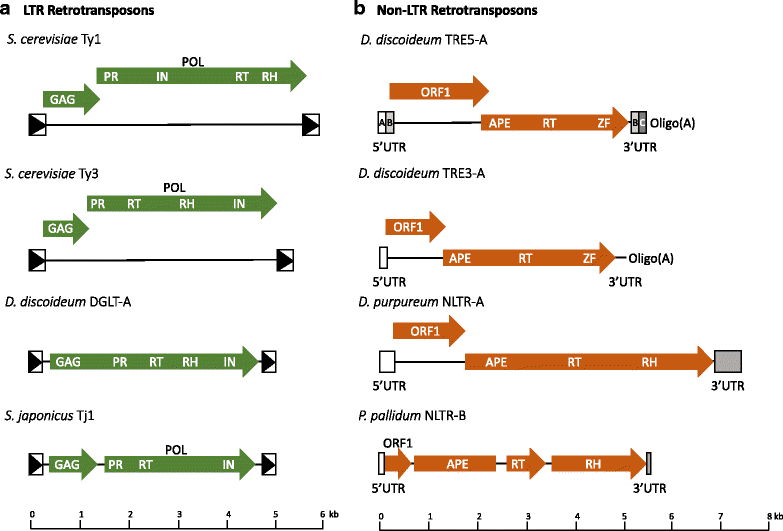
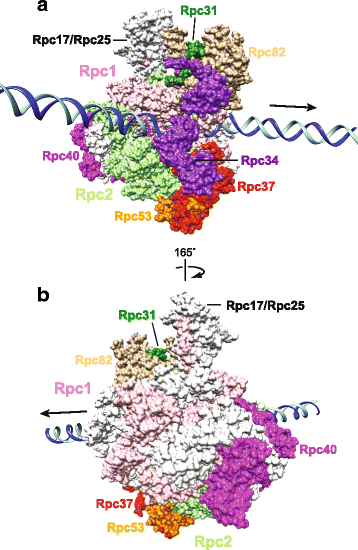
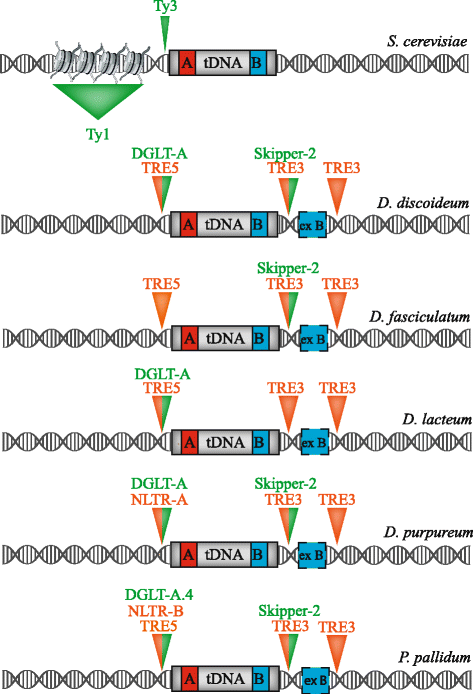
Retrotransposons are genetic elements that are similar in structure and life cycle to retroviruses by replicating via an RNA intermediate and inserting into a host genome. The Saccharomyces cerevisiae (S. cerevisiae) Ty1-5 elements are long terminal repeat (LTR) retrotransposons that are members of the Ty1-copia (Pseudoviridae) or Ty3-gypsy (Metaviridae) families. Four of the five S. cerevisiae Ty elements are inserted into the genome upstream of RNA Polymerase (Pol) III-transcribed genes such as transfer RNA (tRNA) genes. This particular genomic locus provides a safe environment for Ty element insertion without disruption of the host genome and is a targeting strategy used by retrotransposons that insert into compact genomes of hosts such as S. cerevisiae and the social amoeba Dictyostelium. The mechanism by which Ty1 targeting is achieved has been recently solved due to the discovery of an interaction between Ty1 Integrase (IN) and RNA Pol III subunits. We describe the methods used to identify the Ty1-IN interaction with Pol III and the Ty1 targeting consequences if the interaction is perturbed. The details of Ty1 targeting are just beginning to emerge and many unexplored areas remain including consideration of the 3-dimensional shape of genome. We present a variety of other retrotransposon families that insert adjacent to Pol III-transcribed genes and the mechanism by which the host machinery has been hijacked to accomplish this targeting strategy. Finally, we discuss why retrotransposons selected Pol III-transcribed genes as a target during evolution and how retrotransposons have shaped genome architecture.
Keywords: Integrase; RNA polymerase III; Retrotransposon; S. cerevisiae; Ty element; tRNA.
Conflict of interest statement
Not applicable.The authors declare that they have no competing interests.Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
A small targeting domain in Ty1 integrase is sufficient to direct retrotransposon integration upstream of tRNA genes.EMBO J. 2020 Sep 1;39(17):e104337. doi: 10.15252/embj.2019104337. Epub 2020 Jul 17. EMBO J. 2020. PMID: 32677087 Free PMC article.
-
Ty1 Integrase Interacts with RNA Polymerase III-specific Subcomplexes to Promote Insertion of Ty1 Elements Upstream of Polymerase (Pol) III-transcribed Genes.J Biol Chem. 2016 Mar 18;291(12):6396-411. doi: 10.1074/jbc.M115.686840. Epub 2016 Jan 21. J Biol Chem. 2016. PMID: 26797132 Free PMC article.
-
Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence.Genome Res. 1998 May;8(5):464-78. doi: 10.1101/gr.8.5.464. Genome Res. 1998. PMID: 9582191
-
Happy together: the life and times of Ty retrotransposons and their hosts.Cytogenet Genome Res. 2005;110(1-4):70-90. doi: 10.1159/000084940. Cytogenet Genome Res. 2005. PMID: 16093660 Review.
-
Ty3, a Position-specific Retrotransposon in Budding Yeast.Microbiol Spectr. 2015 Apr;3(2):MDNA3-0057-2014. doi: 10.1128/microbiolspec.MDNA3-0057-2014. Microbiol Spectr. 2015. PMID: 26104707 Review.
Cited by
-
On the Track of the Missing tRNA Genes: A Source of Non-Canonical Functions?Front Mol Biosci. 2021 Mar 16;8:643701. doi: 10.3389/fmolb.2021.643701. eCollection 2021. Front Mol Biosci. 2021. PMID: 33796548 Free PMC article. Review.
-
Transposable elements in mammalian chromatin organization.Nat Rev Genet. 2023 Oct;24(10):712-723. doi: 10.1038/s41576-023-00609-6. Epub 2023 Jun 7. Nat Rev Genet. 2023. PMID: 37286742 Review.
-
A small targeting domain in Ty1 integrase is sufficient to direct retrotransposon integration upstream of tRNA genes.EMBO J. 2020 Sep 1;39(17):e104337. doi: 10.15252/embj.2019104337. Epub 2020 Jul 17. EMBO J. 2020. PMID: 32677087 Free PMC article.
-
Locus-specific proteome decoding reveals Fpt1 as a chromatin-associated negative regulator of RNA polymerase III assembly.Mol Cell. 2023 Dec 7;83(23):4205-4221.e9. doi: 10.1016/j.molcel.2023.10.037. Epub 2023 Nov 22. Mol Cell. 2023. PMID: 37995691 Free PMC article.
-
Deciphering recent transposition patterns in plants through comparison of 811 genome assemblies.Plant Biotechnol J. 2025 Apr;23(4):1121-1132. doi: 10.1111/pbi.14570. Epub 2025 Jan 10. Plant Biotechnol J. 2025. PMID: 39791953 Free PMC article.
References
-
- Finnegan DJ. Eukaryotic transposable elements and genome evolution. Trends Genet. 1989;5(4):103–107. - PubMed
-
- Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8(12):973–982. - PubMed
-
- Boeke JD, Garfinkel DJ, Styles CA, Fink GR. Ty elements transpose through an RNA intermediate. Cell. 1985;40(3):491–500. - PubMed
-
- Lesage P, Todeschini AL. Happy together: the life and times of ty retrotransposons and their hosts. Cytogenet Genome Res. 2005;110(1–4):70–90. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

