Metagenome, metatranscriptome, and metaproteome approaches unraveled compositions and functional relationships of microbial communities residing in biogas plants
- PMID: 29713790
- PMCID: PMC5959977
- DOI: 10.1007/s00253-018-8976-7
Metagenome, metatranscriptome, and metaproteome approaches unraveled compositions and functional relationships of microbial communities residing in biogas plants
Abstract
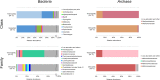
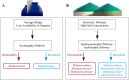
The production of biogas by anaerobic digestion (AD) of agricultural residues, organic wastes, animal excrements, municipal sludge, and energy crops has a firm place in sustainable energy production and bio-economy strategies. Focusing on the microbial community involved in biomass conversion offers the opportunity to control and engineer the biogas process with the objective to optimize its efficiency. Taxonomic profiling of biogas producing communities by means of high-throughput 16S rRNA gene amplicon sequencing provided high-resolution insights into bacterial and archaeal structures of AD assemblages and their linkages to fed substrates and process parameters. Commonly, the bacterial phyla Firmicutes and Bacteroidetes appeared to dominate biogas communities in varying abundances depending on the apparent process conditions. Regarding the community of methanogenic Archaea, their diversity was mainly affected by the nature and composition of the substrates, availability of nutrients and ammonium/ammonia contents, but not by the temperature. It also appeared that a high proportion of 16S rRNA sequences can only be classified on higher taxonomic ranks indicating that many community members and their participation in AD within functional networks are still unknown. Although cultivation-based approaches to isolate microorganisms from biogas fermentation samples yielded hundreds of novel species and strains, this approach intrinsically is limited to the cultivable fraction of the community. To obtain genome sequence information of non-cultivable biogas community members, metagenome sequencing including assembly and binning strategies was highly valuable. Corresponding research has led to the compilation of hundreds of metagenome-assembled genomes (MAGs) frequently representing novel taxa whose metabolism and lifestyle could be reconstructed based on nucleotide sequence information. In contrast to metagenome analyses revealing the genetic potential of microbial communities, metatranscriptome sequencing provided insights into the metabolically active community. Taking advantage of genome sequence information, transcriptional activities were evaluated considering the microorganism's genetic background. Metaproteome studies uncovered enzyme profiles expressed by biogas community members. Enzymes involved in cellulose and hemicellulose decomposition and utilization of other complex biopolymers were identified. Future studies on biogas functional microbial networks will increasingly involve integrated multi-omics analyses evaluating metagenome, transcriptome, proteome, and metabolome datasets.
Keywords: Anaerobic digestion; Biogas microbiome; Biomass conversion; Genome-enabled metatranscriptomics; Integrated omics; Metagenome-assembled genomes; Methanogenesis; Methanogenic Archaea; Sewage digesters; Taxonomic profiling.
Conflict of interest statement
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Figures
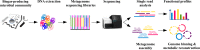
References
-
- Akyol Ç, Aydin S, Ince O, Ince B. A comprehensive microbial insight into single-stage and two-stage anaerobic digestion of oxytetracycline-medicated cattle manure. Chem Eng J. 2016;303:675–684.
-
- Angenent LT, Karim K, Al-Dahhan MH, Wrenn BA, Domíguez-Espinosa R. Production of bioenergy and biochemicals from industrial and agricultural wastewater. Trends Biotechnol. 2004;22:477–485. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

