Transposable elements and polyploid evolution in animals
- PMID: 29715568
- PMCID: PMC5975190
- DOI: 10.1016/j.gde.2018.04.003
Transposable elements and polyploid evolution in animals
Abstract
Polyploidy in animals is much less common than in plants, where it is thought to be pervasive in all higher plant lineages. Recent studies have highlighted the impact of polyploidization and the associated process of diploidy restoration on the evolution and speciation of selected taxonomic groups in the animal kingdom: from vertebrates represented by salmonid fishes and African clawed frogs to invertebrates represented by parasitic root-knot nematodes and bdelloid rotifers. In this review, we focus on the unique and diverse roles that transposable elements may play in these processes, from marking and diversifying subgenome-specific chromosome sets before hybridization, to influencing genome restructuring during rediploidization, to affecting subgenome-specific regulatory evolution, and occasionally providing opportunities for domestication and gene amplification to restore and improve functionality. There is still much to be learned from the future comparative genomic studies of chromosome-sized and haplotype-aware assemblies, and from postgenomic studies elucidating genetic and epigenetic regulatory phenomena across short and long evolutionary distances in the metazoan tree of life.
Copyright © 2018 Elsevier Ltd. All rights reserved.
Conflict of interest statement
None declared.
Figures
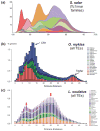
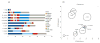
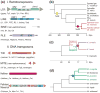
References
-
- White MJD. Animal Cytology and Evolution. 3. Cambridge, UK: Cambridge University Press; 1977.
-
- Otto SP, Whitton J. Polyploid incidence and evolution. Annu Rev Genet. 2000;34:401–437. - PubMed
-
- Soltis PS, Marchant DB, Van de Peer Y, Soltis DE. Polyploidy and genome evolution in plants. Curr Opin Genet Dev. 2015;35:119–125. - PubMed
-
- Jaillon O, Aury JM, Wincker P. “Changing by doubling”, the impact of Whole Genome Duplications in the evolution of eukaryotes. C R Biol. 2009;332:241–253. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

