Lifespan analysis of brain development, gene expression and behavioral phenotypes in the Ts1Cje, Ts65Dn and Dp(16)1/Yey mouse models of Down syndrome
- PMID: 29716957
- PMCID: PMC6031353
- DOI: 10.1242/dmm.031013
Lifespan analysis of brain development, gene expression and behavioral phenotypes in the Ts1Cje, Ts65Dn and Dp(16)1/Yey mouse models of Down syndrome
Abstract
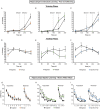
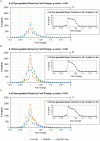
Down syndrome (DS) results from triplication of human chromosome 21. Neuropathological hallmarks of DS include atypical central nervous system development that manifests prenatally and extends throughout life. As a result, individuals with DS exhibit cognitive and motor deficits, and have delays in achieving developmental milestones. To determine whether different mouse models of DS recapitulate the human prenatal and postnatal phenotypes, here, we directly compared brain histogenesis, gene expression and behavior over the lifespan of three cytogenetically distinct mouse models of DS: Ts1Cje, Ts65Dn and Dp(16)1/Yey. Histological data indicated that Ts65Dn mice were the most consistently affected with respect to somatic growth, neurogenesis and brain morphogenesis. Embryonic and adult gene expression results showed that Ts1Cje and Ts65Dn brains had considerably more differentially expressed (DEX) genes compared with Dp(16)1/Yey mice, despite the larger number of triplicated genes in the latter model. In addition, DEX genes showed little overlap in identity and chromosomal distribution in the three models, leading to dissimilarities in affected functional pathways. Perinatal and adult behavioral testing also highlighted differences among the models in their abilities to achieve various developmental milestones and perform hippocampal- and motor-based tasks. Interestingly, Dp(16)1/Yey mice showed no abnormalities in prenatal brain phenotypes, yet they manifested behavioral deficits starting at postnatal day 15 that continued through adulthood. In contrast, Ts1Cje mice showed mildly abnormal embryonic brain phenotypes, but only select behavioral deficits as neonates and adults. Altogether, our data showed widespread and unexpected fundamental differences in behavioral, gene expression and brain development phenotypes between these three mouse models. Our findings illustrate unique limitations of each model when studying aspects of brain development and function in DS. This work helps to inform model selection in future studies investigating how observed neurodevelopmental abnormalities arise, how they contribute to cognitive impairment, and when testing therapeutic molecules to ameliorate the intellectual disability associated with DS.This article has an associated First Person interview with the first author of the paper.
Keywords: Brain development; Developmental disorders; Down syndrome; Dp(16)1/Yey; Lifespan analysis; Ts1Cje; Ts65Dn.
© 2018. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures
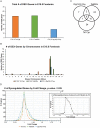
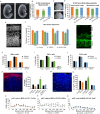
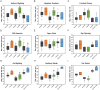

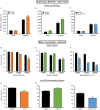

References
-
- Becker L., Mito T., Takashima S. and Onodera K. (1991). Growth and development of the brain in Down syndrome. Prog. Clin. Biol. Res. 373, 133-152. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

