Regulation of Flagellum Biosynthesis in Response to Cell Envelope Stress in Salmonella enterica Serovar Typhimurium
- PMID: 29717015
- PMCID: PMC5930307
- DOI: 10.1128/mBio.00736-17
Regulation of Flagellum Biosynthesis in Response to Cell Envelope Stress in Salmonella enterica Serovar Typhimurium
Abstract
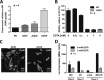
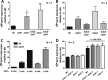
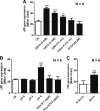
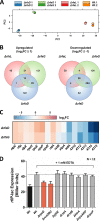
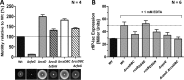
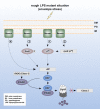
Flagellum-driven motility of Salmonella enterica serovar Typhimurium facilitates host colonization. However, the large extracellular flagellum is also a prime target for the immune system. As consequence, expression of flagella is bistable within a population of Salmonella, resulting in flagellated and nonflagellated subpopulations. This allows the bacteria to maximize fitness in hostile environments. The degenerate EAL domain protein RflP (formerly YdiV) is responsible for the bistable expression of flagella by directing the flagellar master regulatory complex FlhD4C2 with respect to proteolytic degradation. Information concerning the environmental cues controlling expression of rflP and thus about the bistable flagellar biosynthesis remains ambiguous. Here, we demonstrated that RflP responds to cell envelope stress and alterations of outer membrane integrity. Lipopolysaccharide (LPS) truncation mutants of Salmonella Typhimurium exhibited increasing motility defects due to downregulation of flagellar gene expression. Transposon mutagenesis and genetic profiling revealed that σ24 (RpoE) and Rcs phosphorelay-dependent cell envelope stress response systems sense modifications of the lipopolysaccaride, low pH, and activity of the complement system. This subsequently results in activation of RflP expression and degradation of FlhD4C2 via ClpXP. We speculate that the presence of diverse hostile environments inside the host might result in cell envelope damage and would thus trigger the repression of resource-costly and immunogenic flagellum biosynthesis via activation of the cell envelope stress response.IMPORTANCE Pathogenic bacteria such as Salmonella Typhimurium sense and adapt to a multitude of changing and stressful environments during host infection. At the initial stage of gastrointestinal colonization, Salmonella uses flagellum-mediated motility to reach preferred sites of infection. However, the flagellum also constitutes a prime target for the host's immune response. Accordingly, the pathogen needs to determine the spatiotemporal stage of infection and control flagellar biosynthesis in a robust manner. We found that Salmonella uses signals from cell envelope stress-sensing systems to turn off production of flagella. We speculate that downregulation of flagellum synthesis after cell envelope damage in hostile environments aids survival of Salmonella during late stages of infection and provides a means to escape recognition by the immune system.
Copyright © 2018 Spöring et al.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

