OsMADS6 Controls Flower Development by Activating Rice FACTOR OF DNA METHYLATION LIKE1
- PMID: 29717020
- PMCID: PMC6001338
- DOI: 10.1104/pp.18.00017
OsMADS6 Controls Flower Development by Activating Rice FACTOR OF DNA METHYLATION LIKE1
Abstract
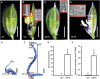
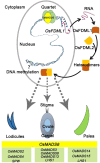
OsMADS6, an ancient AGAMOUS-LIKE6 (AGL6)-like gene, has essential functions in specifying floral organ and meristem identity in rice (Oryza sativa). However, how AGL6 genes control flower development remains largely unknown. In this study, we report that OsMADS6 directly targets FACTOR OF DNA METHYLATION LIKE 1 (OsFDML1), a rice homolog of the SUPPRESSOR OF GENE SILENCING3-like gene FACTOR OF DNA METHYLATION 1 (FDM1) from Arabidopsis (Arabidopsis thaliana). Arabidopsis FDM1 is involved in RNA-directed DNA methylation and OsFDML1 regulates flower development. The expression of OsFDML1 overlaps with that of OsMADS6 in the palea primordia and the ovule, and OsMADS6 directly promotes OsFDML1 expression through binding to regions containing putative CArG motifs within the OsFDML1 promoter during rice spikelet development. Consistent with the phenotypes of osmads6 mutants, the osfdml1 mutants showed floral defects, including altered palea identity with lemma-like shape containing no marginal region of palea, increased numbers of stigmas and fused carpels, and meristem indeterminacy. Moreover, transgenic plants overexpressing OsFDML1 displayed floral defects, such as abnormal paleae. Phylogenetic analysis showed that OsFDML1 homologs exist only in terrestrial plants. In addition, protein-protein interaction assays showed that OsFDML1 interacts with its close paralog OsFDML2, similar to the activity of OsFDML1 homologs in Arabidopsis. These results provide insight into how the ancient AGL6 gene regulates floral development.
© 2018 American Society of Plant Biologists. All rights reserved.
Figures




References
-
- Agrawal GK, Abe K, Yamazaki M, Miyao A, Hirochika H (2005) Conservation of the E-function for floral organ identity in rice revealed by the analysis of tissue culture-induced loss-of-function mutants of the OsMADS1 gene. Plant Mol Biol 59: 125–135 - PubMed
-
- Ambrose BA, Lerner DR, Ciceri P, Padilla CM, Yanofsky MF, Schmidt RJ (2000) Molecular and genetic analyses of the silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol Cell 5: 569–579 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

