Drug-Resistance and Population Structure of Plasmodium falciparum Across the Democratic Republic of Congo Using High-Throughput Molecular Inversion Probes
- PMID: 29718283
- PMCID: PMC6093412
- DOI: 10.1093/infdis/jiy223
Drug-Resistance and Population Structure of Plasmodium falciparum Across the Democratic Republic of Congo Using High-Throughput Molecular Inversion Probes
Abstract
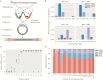
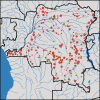
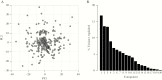
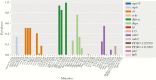
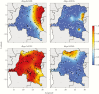
A better understanding of the drivers of the spread of malaria parasites and drug resistance across space and time is needed. These drivers can be elucidated using genetic tools. Here, a novel molecular inversion probe (MIP) panel targeting all major drug-resistance mutations and a set of microsatellites was used to genotype Plasmodium falciparum infections of 552 children from the 2013-2014 Demographic and Health Survey conducted in the Democratic Republic of the Congo (DRC). Microsatellite-based analysis of population structure suggests that parasites within the DRC form a homogeneous population. In contrast, sulfadoxine-resistance markers in dihydropteroate synthase show marked spatial structure with ongoing spread of double and triple mutants compared with 2007. These findings suggest that parasites in the DRC remain panmictic despite rapidly spreading antimalarial-resistance mutations. Moreover, highly multiplexed targeted sequencing using MIPs emerges as a cost-effective method for elucidating pathogen genetics in complex infections in large cohorts.
Figures
References
-
- World Health Organization. World malaria report 2017 http://www.who.int/malaria/publications/world-malaria-report-2017/report.... Accessed 2 February 2018.
-
- Kaufmann C, Briegel H. Flight performance of the malaria vectors Anopheles gambiae and Anopheles atroparvus. J Vector Ecol 2004; 29:140–53. - PubMed
-
- Anderson TJ, Roper C. The origins and spread of antimalarial drug resistance: lessons for policy makers. Acta Trop 2005; 94:269–80. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

