The KDM4A/KDM4C/NF-κB and WDR5 epigenetic cascade regulates the activation of B cells
- PMID: 29718303
- PMCID: PMC6009645
- DOI: 10.1093/nar/gky281
The KDM4A/KDM4C/NF-κB and WDR5 epigenetic cascade regulates the activation of B cells
Abstract
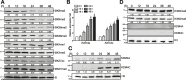
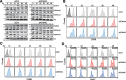
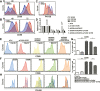
T follicular helper (Tfh) cell-derived signals promote activation and proliferation of antigen-primed B cells. It remains unclear whether epigenetic regulation is involved in the B cell responses to Tfh cell-derived signals. Here, we demonstrate that Tfh cell-mimicking signals induce the expression of histone demethylases KDM4A and KDM4C, and the concomitant global down-regulation of their substrates, H3K9me3/me2, in B cells. Depletion of KDM4A and KDM4C potentiates B cell activation and proliferation in response to Tfh cell-derived signals. ChIP-seq and de novo motif analysis reveals NF-κB p65 as a binding partner of KDM4A and KDM4C. Their co-targeting to Wdr5, a MLL complex member promoting H3K4 methylation, up-regulates cell cycle inhibitors Cdkn2c and Cdkn3. Thus, Tfh cell-derived signals trigger KDM4A/KDM4C - WDR5 - Cdkn2c/Cdkn3 cascade in vitro, an epigenetic mechanism regulating proper proliferation of activated B cells. This pathway is dysregulated in B cells from systemic lupus erythematosus patients and may represent a pathological link.
Figures




References
-
- Kouzarides T. Chromatin modifications and their function. Cell. 2007; 128:693–705. - PubMed
-
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K.. High-resolution profiling of histone methylations in the human genome. Cell. 2007; 129:823–837. - PubMed
-
- Kim K.C., Geng L., Huang S.. Inactivation of a histone methyltransferase by mutations in human cancers. Cancer Res. 2003; 63:7619–7623. - PubMed
-
- Mosammaparast N., Shi Y.. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu. Rev. Biochem. 2010; 79:155–179. - PubMed
-
- Shi Y., Lan F., Matson C., Mulligan P., Whetstine J.R., Cole P.A., Casero R.A., Shi Y.. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004; 119:941–953. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

