Molecular Landscape of ERBB2/ERBB3 Mutated Colorectal Cancer
- PMID: 29718453
- PMCID: PMC6292791
- DOI: 10.1093/jnci/djy067
Molecular Landscape of ERBB2/ERBB3 Mutated Colorectal Cancer
Abstract
Background: Despite growing therapeutic relevance of ERBB2 amplifications in colorectal cancer (CRC), little is known about ERBB2/ERBB3 mutations. We aimed to characterize these subsets of CRC.
Methods: We performed a retrospective analysis of 419 CRC patients from MD Anderson (MDACC) and 619 patients from the Nurses' Health Study (NHS)/Health Professionals Follow-Up Study (HPFS) with tissue sequencing, clinicopathologic, mutational, and consensus molecular subtype (CMS) profiles of ERBB2/ERBB3 mutant patients. A third cohort of 1623 CRC patients with ctDNA assays characterized the ctDNA profile of ERBB2 mutants. All statistical tests were two-sided.
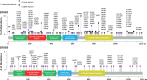
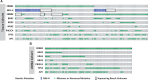
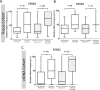
Results: ERBB2 mutations occurred in 4.1% (95% confidence interval [CI] = 2.4% to 6.4%), 5.8% (95% CI = 4.1% to 8.0%), and 5.1% (95% CI = 4.0% to 6.2%) of MDACC, NHS/HPFS, and ctDNA patients, respectively. ERBB3 mutations occurred in 5.7% (95% CI = 3.7% to 8.4%, 95% CI = 4.0% to 7.8%) of patients in both tissue cohorts. Age, stage, and tumor location were not associated with either mutation. Microsatellite instability (MSI) was associated with ERBB2 (odds ratio [OR] = 5.98, 95% CI = 2.47 to 14.49, P < .001; OR = 5.13, 95% CI = 2.38 to 11.05, P < .001) and ERBB3 mutations (OR = 3.48, 95% CI = 1.51 to 8.02, P = .002; OR = 3.40, 95% CI = 1.05 to 10.96, P = .03) in both tissue cohorts. Neither gene was associated with TP53, APC, KRAS, NRAS, or BRAF mutations in tissue. However, PIK3CA mutations were strongly associated with ERBB2 mutations in all three cohorts (OR = 3.68, 95% CI = 1.83 to 7.41, P = .001; OR = 2.25, 95% CI = 1.11 to 4.58, P = .02; OR = 2.11, 95% CI = 1.25 to 3.58, P = .004) and ERBB3 mutations in the MDACC cohort (OR = 13.26, 95% CI = 5.27 to 33.33, P < .001). ERBB2 (P = 0.08) and ERBB3 (P = .008) mutations were associated with CMS1 subtype. ERBB2 (hazard ratio [HR] = 1.82, 95% CI = 1.23 to 4.03, P = .009), but not ERBB3 (HR = 0.88, 95% CI = 0.45 to 1.73, P = .73), mutations were associated with worse overall survival.
Conclusions: MSI and PIK3CA mutations are associated with ERBB2/ERBB3 mutations. Co-occurring PIK3CA mutations may represent a second hit to oncogenic signaling that needs consideration when targeting ERBB2/ERBB3.
Figures

References
-
- Bertotti A, Migliardi G, Galimi F, et al. A molecularly annotated platform of patient-derived xenografts identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov. 2011;16:508–523. - PubMed
-
- Hurwitz H, Hainsworth JD, Swanton C, et al. Targeted therapy for gastrointestinaI (GI) tumors based on molecular profiles: Early results from MyPathway, an open - label phase IIa basket study in patients with advanced solid Abstracts by Herbert Hurwitz. J Clin Oncol. 2016;34(Supplement 4S):Abstract 653.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

