Proteomic alterations in early stage cervical cancer
- PMID: 29719595
- PMCID: PMC5915062
- DOI: 10.18632/oncotarget.24773
Proteomic alterations in early stage cervical cancer
Abstract
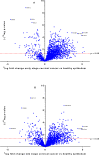
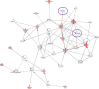
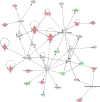
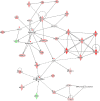
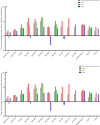
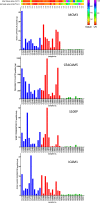
Laser capture microdissection (LCM) allows the capture of cell types or well-defined structures in tissue. We compared in a semi-quantitative way the proteomes from an equivalent of 8,000 tumor cells from patients with squamous cell cervical cancer (SCC, n = 22) with healthy epithelial and stromal cells obtained from normal cervical tissue (n = 13). Proteins were enzymatically digested into peptides which were measured by high-resolution mass spectrometry and analyzed by "all-or-nothing" analysis, Bonferroni, and Benjamini-Hochberg correction for multiple testing. By comparing LCM cell type preparations, 31 proteins were exclusively found in early stage cervical cancer (n = 11) when compared with healthy epithelium and stroma, based on criteria that address specificity in a restrictive "all-or-nothing" way. By Bonferroni correction for multiple testing, 30 proteins were significantly up-regulated between early stage cervical cancer and healthy control, including six members of the MCM protein family. MCM proteins are involved in DNA repair and expected to be participating in the early stage of cancer. After a less stringent Benjamini-Hochberg correction for multiple testing, we found that the abundances of 319 proteins were significantly different between early stage cervical cancer and healthy controls. Four proteins were confirmed in digests of whole tissue lysates by Parallel Reaction Monitoring (PRM). Ingenuity Pathway Analysis using correction for multiple testing by permutation resulted in two networks that were differentially regulated in early stage cervical cancer compared with healthy tissue. From these networks, we learned that specific tumor mechanisms become effective during the early stage of cervical cancer.
Keywords: LCM; PRM; biomarker; cervical cancer; proteomics.
Conflict of interest statement
CONFLICTS OF INTEREST The authors report no conflicts of interest.
Figures



Similar articles
-
Proteomic profiling of human islets collected from frozen pancreata using laser capture microdissection.J Proteomics. 2017 Jan 6;150:149-159. doi: 10.1016/j.jprot.2016.09.002. Epub 2016 Sep 13. J Proteomics. 2017. PMID: 27620696 Free PMC article.
-
Reproducibility of protein identification of selected cell types in Barrett's esophagus analyzed by combining laser-capture microdissection and mass spectrometry.J Proteome Res. 2011 Jan 7;10(1):288-98. doi: 10.1021/pr100709b. Epub 2010 Dec 3. J Proteome Res. 2011. PMID: 21053923
-
Proteomic analysis of high-grade dysplastic cervical cells obtained from ThinPrep slides using laser capture microdissection and mass spectrometry.J Proteome Res. 2007 Nov;6(11):4256-68. doi: 10.1021/pr070319j. Epub 2007 Sep 29. J Proteome Res. 2007. PMID: 17902640
-
Quantitative proteomic analysis of microdissected oral epithelium for cancer biomarker discovery.Oral Oncol. 2015 Nov;51(11):1011-1019. doi: 10.1016/j.oraloncology.2015.08.008. Epub 2015 Aug 29. Oral Oncol. 2015. PMID: 26321370
-
Combining laser capture microdissection and proteomics: methodologies and clinical applications.Proteomics Clin Appl. 2010 Feb;4(2):116-23. doi: 10.1002/prca.200900138. Epub 2009 Nov 19. Proteomics Clin Appl. 2010. PMID: 21137037 Review.
Cited by
-
Inflammatory cytokines and a diverse cervicovaginal microbiota associate with cervical dysplasia in a cohort of Hispanics living in Puerto Rico.PLoS One. 2023 Dec 8;18(12):e0284673. doi: 10.1371/journal.pone.0284673. eCollection 2023. PLoS One. 2023. PMID: 38064478 Free PMC article.
-
Unveiling diagnostic and therapeutic strategies for cervical cancer: biomarker discovery through proteomics approaches and exploring the role of cervical cancer stem cells.Front Oncol. 2024 Jan 24;13:1277772. doi: 10.3389/fonc.2023.1277772. eCollection 2023. Front Oncol. 2024. PMID: 38328436 Free PMC article. Review.
-
Novel CSF biomarkers in genetic frontotemporal dementia identified by proteomics.Ann Clin Transl Neurol. 2019 Mar 7;6(4):698-707. doi: 10.1002/acn3.745. eCollection 2019 Apr. Ann Clin Transl Neurol. 2019. PMID: 31019994 Free PMC article.
-
Human Papillomavirus 16 E6 and E7 Oncoproteins Alter the Abundance of Proteins Associated with DNA Damage Response, Immune Signaling and Epidermal Differentiation.Viruses. 2022 Aug 12;14(8):1764. doi: 10.3390/v14081764. Viruses. 2022. PMID: 36016386 Free PMC article.
-
Silencing DTX3L Inhibits the Progression of Cervical Carcinoma by Regulating PI3K/AKT/mTOR Signaling Pathway.Int J Mol Sci. 2023 Jan 3;24(1):861. doi: 10.3390/ijms24010861. Int J Mol Sci. 2023. PMID: 36614304 Free PMC article.
References
-
- Franco EL, Schlecht NF, Saslow D. The epidemiology of cervical cancer. Cancer journal. 2003;9:348–359. - PubMed
-
- Soerjomataram I, Lortet-Tieulent J, Parkin DM, Ferlay J, Mathers C, Forman D, Bray F. Global burden of cancer in 2008: a systematic analysis of disability-adjusted life-years in 12 world regions. Lancet. 2012;380:1840–1850. - PubMed
-
- Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

