Crystal structure of the human 4-1BB/4-1BBL complex
- PMID: 29720399
- PMCID: PMC6016478
- DOI: 10.1074/jbc.RA118.002803
Crystal structure of the human 4-1BB/4-1BBL complex
Abstract
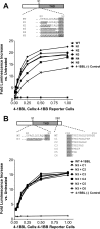
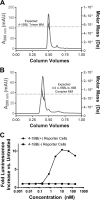
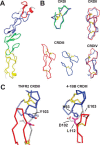
4-1BBL is a member of the tumor necrosis factor (TNF) superfamily and is the ligand for the TNFR superfamily receptor, 4-1BB. 4-1BB plays an immunomodulatory role in T cells and NK cells, and agonists of this receptor have garnered strong attention as potential immunotherapy agents. Broadly speaking, the structural features of TNF superfamily members, their receptors, and ligand-receptor complexes are similar. However, a published crystal structure of human 4-1BBL suggests that it may be unique in this regard, exhibiting a three-bladed propeller-like trimer assembly that is distinctly different from that observed in other family members. This unusual structure also suggests that the human 4-1BB/4-1BBL complex may be structurally unique within the TNF/TNFR superfamily, but to date no structural data have been reported. Here we report the crystal structure of the human 4-1BB/4-1BBL complex at 2.4-Å resolution. In this structure, 4-1BBL does not adopt the unusual trimer assembly previously reported, but instead forms a canonical bell-shaped trimer typical of other TNF superfamily members. The structure of 4-1BB is also largely canonical as is the 4-1BB/4-1BBL complex. Mutational data support the 4-1BBL structure reported here as being biologically relevant, suggesting that the previously reported structure is not. Together, the data presented here offer insight into structure/function relationships in the 4-1BB/4-1BBL system and improve our structural understanding of the TNF/TNFR superfamily more broadly.
Keywords: T-cell biology; crystal structure; protein structure; protein-protein interaction; structural biology; structure-function; tumor necrosis factor (TNF).
© 2018 Gilbreth et al.
Conflict of interest statement
All authors are or have been employees of and may hold financial interests in MedImmune LLC.
Figures



References
-
- Sanmamed M. F., Pastor F., Rodriguez A., Perez-Gracia J. L., Rodriguez-Ruiz M. E., Jure-Kunkel M., and Melero I. (2015) Agonists of co-stimulation in cancer immunotherapy directed against CD137, OX40, GITR, CD27, CD28, and ICOS. Semin. Oncol. 42, 640–655 10.1053/j.seminoncol.2015.05.014 - DOI - PubMed
-
- Chester C., Sanmamed M. F., Wang J., and Melero I. (2018) Immunotherapy targeting 4-1BB: mechanistic rationale, clinical results, and future strategies. Blood 131, 49–57 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

