The nanos1 gene was duplicated in early Vertebrates and the two paralogs show different gonadal expression profiles in a shark
- PMID: 29720681
- PMCID: PMC5932020
- DOI: 10.1038/s41598-018-24643-1
The nanos1 gene was duplicated in early Vertebrates and the two paralogs show different gonadal expression profiles in a shark
Erratum in
-
Author Correction: The nanos1 gene was duplicated in early Vertebrates and the two paralogs show different gonadal expression profiles in a shark.Sci Rep. 2024 Mar 18;14(1):6516. doi: 10.1038/s41598-024-56271-3. Sci Rep. 2024. PMID: 38499596 Free PMC article. No abstract available.
Abstract
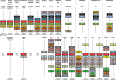
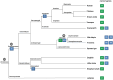
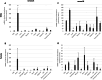
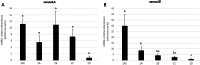
Nanos are RNA-binding proteins playing crucial roles in germ cell development and maintenance. Based on phylogenetic and synteny analyses, this study reveals that nanos1 gene has undergone multiple duplications and gene copies losses in Vertebrates. Chondrichthyan species display two nanos1 genes (named nanos1A/1B), which were both retrieved in some Osteichthyes at basal positions in Sarcopterygii and Actinopterygii lineages. In contrast, Teleosts have lost nanos1A but duplicated nanos1B leading to the emergence of two ohnologs (nanos1Ba/1Bb), whereas Tetrapods have lost nanos1B gene. The two successive nanos gene duplications may result from the second and third whole genome duplication events at the basis of Vertebrates and Teleosts respectively. The expression profiles of nanos1A and nanos1B paralogs were characterized in the dogfish, Scyliorhinus canicula. Nanos1A was strongly expressed in brain and also localized in all germ cell types in the polarized testis. In contrast, nanos1B was detected in testis with the highest expression in the germinative zone. In addition, Nanos1B protein was predominantly located in the nuclei of male germinal cells. In the ovary, both paralogs were detected in germinal and somatic cells. Our study opens new perspectives concerning the complex evolution of nanos1 paralogs and their potential distinct roles in Vertebrates gonads.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Expression and syntenic analyses of four nanos genes in medaka.Zoolog Sci. 2009 Feb;26(2):112-8. doi: 10.2108/zsj.26.112. Zoolog Sci. 2009. PMID: 19341327
-
Ancient Duplications and Expression Divergence in the Globin Gene Superfamily of Vertebrates: Insights from the Elephant Shark Genome and Transcriptome.Mol Biol Evol. 2015 Jul;32(7):1684-94. doi: 10.1093/molbev/msv054. Epub 2015 Mar 4. Mol Biol Evol. 2015. PMID: 25743544 Free PMC article.
-
Basal teleosts provide new insights into the evolutionary history of teleost-duplicated aromatase.Gen Comp Endocrinol. 2020 May 15;291:113395. doi: 10.1016/j.ygcen.2020.113395. Epub 2020 Jan 23. Gen Comp Endocrinol. 2020. PMID: 31981691
-
New Insights Into the Evolutionary History of Melatonin Receptors in Vertebrates, With Particular Focus on Teleosts.Front Endocrinol (Lausanne). 2020 Sep 24;11:538196. doi: 10.3389/fendo.2020.538196. eCollection 2020. Front Endocrinol (Lausanne). 2020. PMID: 33071966 Free PMC article. Review.
-
Evolution of endothelin receptors in vertebrates.Gen Comp Endocrinol. 2014 Dec 1;209:21-34. doi: 10.1016/j.ygcen.2014.06.028. Epub 2014 Jul 8. Gen Comp Endocrinol. 2014. PMID: 25010382 Review.
Cited by
-
Emerging Roles of NANOS RNA-Binding Proteins in Cancer.Int J Mol Sci. 2022 Aug 20;23(16):9408. doi: 10.3390/ijms23169408. Int J Mol Sci. 2022. PMID: 36012673 Free PMC article.
-
Characterization of ddx4 and dnd Homologs in Snakeskin Gourami (Trichopodus pectoralis) and Their Expression Levels during Larval Development and in Gonads of Males and Females.Animals (Basel). 2022 Dec 4;12(23):3415. doi: 10.3390/ani12233415. Animals (Basel). 2022. PMID: 36496935 Free PMC article.
-
Molecular Cloning and Sexually Dimorphic Expression Analysis of nanos2 in the Sea Urchin, Mesocentrotus nudus.Int J Mol Sci. 2019 Jun 1;20(11):2705. doi: 10.3390/ijms20112705. Int J Mol Sci. 2019. PMID: 31159444 Free PMC article.
-
Primordial Germ Cell Specification in Vertebrate Embryos: Phylogenetic Distribution and Conserved Molecular Features of Preformation and Induction.Front Cell Dev Biol. 2021 Sep 16;9:730332. doi: 10.3389/fcell.2021.730332. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34604230 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

