Wheat miRNA TaemiR408 Acts as an Essential Mediator in Plant Tolerance to Pi Deprivation and Salt Stress via Modulating Stress-Associated Physiological Processes
- PMID: 29720988
- PMCID: PMC5916090
- DOI: 10.3389/fpls.2018.00499
Wheat miRNA TaemiR408 Acts as an Essential Mediator in Plant Tolerance to Pi Deprivation and Salt Stress via Modulating Stress-Associated Physiological Processes
Abstract
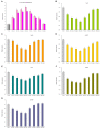
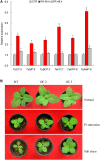
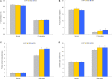
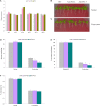
MicroRNAs (miRNA) families act as critical regulators for plant growth, development, and responses to abiotic stresses. In this study, we characterized TaemiR408, a miRNA family member of wheat (Triticum aestivum), for the role in mediating plant responses to Pi starvation and salt stress. TaemiR408 targets six genes that encode proteins involving biochemical metabolism, microtubule organization, and signaling transduction. 5'- and 3'-RACE analyses confirmed the mRNA cleavage of target genes mediated by this wheat miRNA. TaemiR408 showed induced expression patterns upon Pi starvation and salt stress and whose upregulated expression was gradually repressed by the normal recovery treatments. The target genes of TaemiR408 exhibited reverse expression patterns to this miRNA, whose transcripts were downregulated under Pi starvation and salt stress and the reduced expression was recovered by the followed normal condition. These results suggest the regulation of the target genes under TaemiR408 through a cleavage mechanism. Tobacco lines with TaemiR408 overexpression exhibited enhanced stress tolerance, showing improved phenotype, biomass, and photosynthesis behavior compared with wild type under both Pi starvation and salt treatments, which closely associate increased P accumulation upon Pi deprivation and elevated osmolytes under salt stress, respectively. Phosphate transporter (PT) gene NtPT2 displays upregulated transcripts in the Pi-deprived TaemiR408 overexpressors; knockdown of this PT gene reduces Pi acquisition under low-Pi stress, confirming its role in improving plant Pi taken up. Likewise, NtPYL2 and NtSAPK3, genes encoding abscisic acid (ABA) receptor and SnRK2 protein, respectively, exhibited upregulated transcripts in salt-challenged TaemiR408 overexpressors; knockdown of them caused deteriorated growth and lowered osmolytes amounts of plants upon salt treatment. Thus, TaemiR408 is crucial for plant adaptations to Pi starvation and salt stress through regulating Pi acquisition under low-Pi stress and remodel ABA signaling pathway and osmoprotects biosynthesis under salt stress.
Keywords: Pi acquisition; Pi starvation; abiscisic acid signaling; miRNA member; plant growth; salt stress; wheat (Triticum aestivum L.).
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

