Transposable Element Genomic Fissuring in Pyrenophora teres Is Associated With Genome Expansion and Dynamics of Host-Pathogen Genetic Interactions
- PMID: 29720997
- PMCID: PMC5915480
- DOI: 10.3389/fgene.2018.00130
Transposable Element Genomic Fissuring in Pyrenophora teres Is Associated With Genome Expansion and Dynamics of Host-Pathogen Genetic Interactions
Abstract
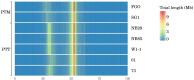
Pyrenophora teres, P. teres f. teres (PTT) and P. teres f. maculata (PTM) cause significant diseases in barley, but little is known about the large-scale genomic differences that may distinguish the two forms. Comprehensive genome assemblies were constructed from long DNA reads, optical and genetic maps. As repeat masking in fungal genomes influences the final gene annotations, an accurate and reproducible pipeline was developed to ensure comparability between isolates. The genomes of the two forms are highly collinear, each composed of 12 chromosomes. Genome evolution in P. teres is characterized by genome fissuring through the insertion and expansion of transposable elements (TEs), a process that isolates blocks of genic sequence. The phenomenon is particularly pronounced in PTT, which has a larger, more repetitive genome than PTM and more recent transposon activity measured by the frequency and size of genome fissures. PTT has a longer cultivated host association and, notably, a greater range of host-pathogen genetic interactions compared to other Pyrenophora spp., a property which associates better with genome size than pathogen lifestyle. The two forms possess similar complements of TE families with Tc1/Mariner and LINE-like Tad-1 elements more abundant in PTT. Tad-1 was only detectable as vestigial fragments in PTM and, within the forms, differences in genome sizes and the presence and absence of several TE families indicated recent lineage invasions. Gene differences between P. teres forms are mainly associated with gene-sparse regions near or within TE-rich regions, with many genes possessing characteristics of fungal effectors. Instances of gene interruption by transposons resulting in pseudogenization were detected in PTT. In addition, both forms have a large complement of secondary metabolite gene clusters indicating significant capacity to produce an array of different molecules. This study provides genomic resources for functional genetics to help dissect factors underlying the host-pathogen interactions.
Keywords: Hordeum vulgare; barley net blotch; optical mapping; repeat annotation; single molecule sequencing DNA; synteny; transposable element.
Figures



References
-
- Akhavan A., Turkington T. K., Kebede B., Xi K., Kumar K., Tekauz A., et al. (2016). Genetic structure of Pyrenophora teres f. teres and P. teres f. maculata populations from western Canada. Eur. J. Plant Pathol. 146 325–335. 10.1007/s10658-016-0919-5 - DOI
-
- Bach E., Christensen S., Dalgaard L., Larsen P. O., Olsen C. E., Smedegård-Petersen V. (1979). Structures, properties and relationship to the aspergillomarasmines of toxins produced by Pyrenophora teres. Physiol. Plant Pathol. 14 41–46. 10.1016/0048-4059(79)90023-7 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

