Integration of genomics and metabolomics for prioritization of rare disease variants: a 2018 literature review
- PMID: 29721916
- PMCID: PMC5959954
- DOI: 10.1007/s10545-018-0139-6
Integration of genomics and metabolomics for prioritization of rare disease variants: a 2018 literature review
Abstract
Many inborn errors of metabolism (IEMs) are amenable to treatment; therefore, early diagnosis and treatment is imperative. Despite recent advances, the genetic basis of many metabolic phenotypes remains unknown. For discovery purposes, whole exome sequencing (WES) variant prioritization coupled with clinical and bioinformatics expertise is the primary method used to identify novel disease-causing variants; however, causation is often difficult to establish due to the number of plausible variants. Integrated analysis of untargeted metabolomics (UM) and WES or whole genome sequencing (WGS) data is a promising systematic approach for identifying disease-causing variants. In this review, we provide a literature-based overview of UM methods utilizing liquid chromatography mass spectrometry (LC-MS), and assess approaches to integrating WES/WGS and LC-MS UM data for the discovery and prioritization of variants causing IEMs. To embed this integrated -omics approach in the clinic, expansion of gene-metabolite annotations and metabolomic feature-to-metabolite mapping methods are needed.
Keywords: Genomics; Inborn errors of metabolism; Metabolomics; Omic integration; Variant prioritization.
Conflict of interest statement
None.
Figures
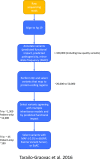
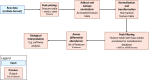

References
-
- Abella L, Steindl K, Simmons L et al (2016) A combined metabolic-genetic approach to early-onset epileptic encephalopathies: results from a Swiss study cohort. Neuropediatrics. doi: 10.1055/s-0036-1583731
-
- Alonso-herranz JGV, Barbas C, Grace E (2015) Controlling the quality of metabolomics data : new strategies to get the best out of the QC sample. Metabolomics 518–528. doi: 10.1007/s11306-014-0712-4
-
- Blau N, Gibson MK, Marinus D, Dionisi-Vici C. IEMBASE, a knowledgebase of inborn errors of metabolism. Mol Genet Metab. 2014;111:296.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

