Whole-Exome Sequencing Identifies Novel Variants that Co-segregates with Autosomal Recessive Retinal Degeneration in a Pakistani Pedigree
- PMID: 29721947
- PMCID: PMC12123434
- DOI: 10.1007/978-3-319-75402-4_27
Whole-Exome Sequencing Identifies Novel Variants that Co-segregates with Autosomal Recessive Retinal Degeneration in a Pakistani Pedigree
Abstract
Purpose: To identify the molecular basis of inherited retinal degeneration (IRD) in a familial case of Pakistani origin using whole-exome sequencing.
Methods: A thorough ophthalmic examination was completed, and genomic DNA was extracted using standard protocols. Whole exome(s) were captured with Agilent V5 + UTRs probes and sequenced on Illumina HiSeq genome analyzer. The exomeSuite software was used to filter variants, and the candidate causal variants were prioritized, examining their allele frequency and PolyPhen2, SIFT, and MutationTaster predictions. Sanger dideoxy sequencing was performed to confirm the segregation with disease phenotype and absence in ethnicity-matched control chromosomes.
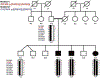
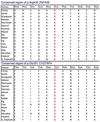
Results: Ophthalmic examination confirmed retinal degeneration in all affected individuals that segregated as an autosomal recessive trait in the family. Whole-exome sequencing identified two homozygous missense variants: c.1304G > A; p.Arg435Gln in ZNF408 (NM_024741) and c.902G > A; p.Gly301Asp in C1QTNF4 (NM_031909). Both variants segregated with the retinal phenotype in the PKRD320 and were absent in ethnically matched control chromosomes.
Conclusion: Whole-exome sequencing coupled with bioinformatics analysis identified potential novel variants that might be responsible for IRD.
Keywords: C1QTNF4; Novel variants; Retinal degeneration; Whole-exome sequencing; ZNF408.
Figures
Similar articles
-
Investigating the Molecular Basis of Retinal Degeneration in a Familial Cohort of Pakistani Decent by Exome Sequencing.PLoS One. 2015 Sep 9;10(9):e0136561. doi: 10.1371/journal.pone.0136561. eCollection 2015. PLoS One. 2015. PMID: 26352687 Free PMC article.
-
Deciphering the genetic architecture and ethnographic distribution of IRD in three ethnic populations by whole genome sequence analysis.PLoS Genet. 2021 Oct 18;17(10):e1009848. doi: 10.1371/journal.pgen.1009848. eCollection 2021 Oct. PLoS Genet. 2021. PMID: 34662339 Free PMC article.
-
Genetic analysis of 10 pedigrees with inherited retinal degeneration by exome sequencing and phenotype-genotype association.Physiol Genomics. 2017 Apr 1;49(4):216-229. doi: 10.1152/physiolgenomics.00096.2016. Epub 2017 Jan 27. Physiol Genomics. 2017. PMID: 28130426 Free PMC article.
-
Identification of a novel mutation in the CDHR1 gene in a family with recessive retinal degeneration.Arch Ophthalmol. 2012 Oct;130(10):1301-8. doi: 10.1001/archophthalmol.2012.1906. Arch Ophthalmol. 2012. PMID: 23044944 Free PMC article.
-
A mutation in IFT43 causes non-syndromic recessive retinal degeneration.Hum Mol Genet. 2017 Dec 1;26(23):4741-4751. doi: 10.1093/hmg/ddx356. Hum Mol Genet. 2017. PMID: 28973684 Free PMC article.
Cited by
-
State of the Art on Inherited Retinal Dystrophies: Management and Molecular Genetics.J Clin Med. 2025 May 18;14(10):3526. doi: 10.3390/jcm14103526. J Clin Med. 2025. PMID: 40429522 Free PMC article. Review.
References
-
- Avila-Fernandez A, Perez-Carro R, Corton M et al. (2015) Whole-exome sequencing reveals ZNF408 as a new gene associated with autosomal recessive retinitis pigmentosa with vitreal alterations. Hum Mol Genet 24:4037–4048 - PubMed
-
- Ayyagari R, Kakuk LE, Bingham EL et al. (2000) Spectrum of color gene deletions and phenotype in patients with blue cone monochromacy. Hum Genet 107:75–82 - PubMed
-
- Ayyagari R, Mandal MN, Karoukis AJ et al. (2005) Late-onset macular degeneration and long anterior lens zonules result from a CTRP5 gene mutation. Invest Ophthalmol Vis Sci 46:3363–3371 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

