Histone acetylation maps in aged mice developmentally exposed to lead: epigenetic drift and Alzheimer-related genes
- PMID: 29722544
- PMCID: PMC5992572
- DOI: 10.2217/epi-2017-0143
Histone acetylation maps in aged mice developmentally exposed to lead: epigenetic drift and Alzheimer-related genes
Abstract
Aim: Early life exposure to lead (Pb) has been shown to increase late life biomarkers involved in Alzheimer's disease (AD) pathology. Here, we tested the hypothesis that latent over expression of AD-related genes may be regulated through histone activation pathways.
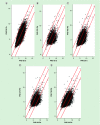
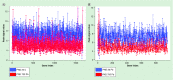
Methods: Chromatin immunoprecipitation sequencing was used to map the histone activation mark (H3K9Ac) to the mouse genome in developmentally Pb exposed mice on postnatal days 20, 270 and 700.
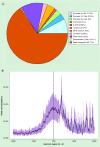
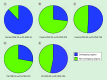
Results: Exposure to Pb resulted in a global downregulation of H3K9Ac across the lifespan; except in genes associated with the Alzheimer pathway.
Discussion: Early life exposure to Pb results in an epigenetic drift in H3K9Ac consistent with latent global gene repression. Alzheimer-related genes do not follow this trend.
Keywords: Alzheimer's disease; epigenetics; histone acetylation; lead (Pb).
Conflict of interest statement
This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences, and by grant 5RO1ES015867-03. This research is based in part upon work conducted using the Rhode Island IDeA Network for Excellence in Biomedical Research Bioinformatics Core which is supported by the NIH under grant 2P20GM103430. JM Gaspar and RP Hart were supported by NIH grants 1R01AT009152 to A-N Kong of Rutgers University and 1R01ES026057 and R21DA035594 to RP Hart. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Figures
References
-
- Portha B, Fournier A, Kioon MD, Mezger V, Movassat J. Early environmental factors, alteration of epigenetic marks and metabolic disease susceptibility. Biochimie. 2014;97:1–15. - PubMed
-
- Tanner CM, Goldman SM, Ross GW, Grate SJ. The disease intersection of susceptibility and exposure: chemical exposures and neurodegenerative disease risk. Alzheimers Dement. 2014;10(3 Suppl.):S213–S225. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
