Potential genetic modifiers of disease risk and age at onset in patients with frontotemporal lobar degeneration and GRN mutations: a genome-wide association study
- PMID: 29724592
- PMCID: PMC6237181
- DOI: 10.1016/S1474-4422(18)30126-1
Potential genetic modifiers of disease risk and age at onset in patients with frontotemporal lobar degeneration and GRN mutations: a genome-wide association study
Abstract
Background: Loss-of-function mutations in GRN cause frontotemporal lobar degeneration (FTLD). Patients with GRN mutations present with a uniform subtype of TAR DNA-binding protein 43 (TDP-43) pathology at autopsy (FTLD-TDP type A); however, age at onset and clinical presentation are variable, even within families. We aimed to identify potential genetic modifiers of disease onset and disease risk in GRN mutation carriers.
Methods: The study was done in three stages: a discovery stage, a replication stage, and a meta-analysis of the discovery and replication data. In the discovery stage, genome-wide logistic and linear regression analyses were done to test the association of genetic variants with disease risk (case or control status) and age at onset in patients with a GRN mutation and controls free of neurodegenerative disorders. Suggestive loci (p<1 × 10-5) were genotyped in a replication cohort of patients and controls, followed by a meta-analysis. The effect of genome-wide significant variants at the GFRA2 locus on expression of GFRA2 was assessed using mRNA expression studies in cerebellar tissue samples from the Mayo Clinic brain bank. The effect of the GFRA2 locus on progranulin concentrations was studied using previously generated ELISA-based expression data. Co-immunoprecipitation experiments in HEK293T cells were done to test for a direct interaction between GFRA2 and progranulin.
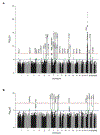
Findings: Individuals were enrolled in the current study between Sept 16, 2014, and Oct 5, 2017. After quality control measures, statistical analyses in the discovery stage included 382 unrelated symptomatic GRN mutation carriers and 1146 controls free of neurodegenerative disorders collected from 34 research centres located in the USA, Canada, Australia, and Europe. In the replication stage, 210 patients (67 symptomatic GRN mutation carriers and 143 patients with FTLD without GRN mutations pathologically confirmed as FTLD-TDP type A) and 1798 controls free of neurodegenerative diseases were recruited from 26 sites, 20 of which overlapped with the discovery stage. No genome-wide significant association with age at onset was identified in the discovery or replication stages, or in the meta-analysis. However, in the case-control analysis, we replicated the previously reported TMEM106B association (rs1990622 meta-analysis odds ratio [OR] 0·54, 95% CI 0·46-0·63; p=3·54 × 10-16), and identified a novel genome-wide significant locus at GFRA2 on chromosome 8p21.3 associated with disease risk (rs36196656 meta-analysis OR 1·49, 95% CI 1·30-1·71; p=1·58 × 10-8). Expression analyses showed that the risk-associated allele at rs36196656 decreased GFRA2 mRNA concentrations in cerebellar tissue (p=0·04). No effect of rs36196656 on plasma and CSF progranulin concentrations was detected by ELISA; however, co-immunoprecipitation experiments in HEK293T cells did suggest a direct binding of progranulin and GFRA2.
Interpretation: TMEM106B-related and GFRA2-related pathways might be future targets for treatments for FTLD, but the biological interaction between progranulin and these potential disease modifiers requires further study. TMEM106B and GFRA2 might also provide opportunities to select and stratify patients for future clinical trials and, when more is known about their potential effects, to inform genetic counselling, especially for asymptomatic individuals.
Funding: National Institute on Aging, National Institute of Neurological Disorders and Stroke, Canadian Institutes of Health Research, Italian Ministry of Health, UK National Institute for Health Research, National Health and Medical Research Council of Australia, and the French National Research Agency.
Copyright © 2018 Elsevier Ltd. All rights reserved.
Figures

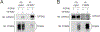
Comment in
-
GFRA2 in GRN-related frontotemporal lobar degeneration.Lancet Neurol. 2018 Jun;17(6):488-489. doi: 10.1016/S1474-4422(18)30171-6. Epub 2018 Apr 30. Lancet Neurol. 2018. PMID: 29724593 No abstract available.
References
-
- Graff-Radford NR, Woodruff BK. Frontotemporal dementia. Semin Neurol 2007; 27(1): 48–57. - PubMed
-
- Neumann M, Sampathu DM, Kwong LK, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 2006; 314(5796): 130–3. - PubMed
-
- Baker M, Mackenzie IR, Pickering-Brown SM, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 2006; 442(7105): 916–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DC008552/DC/NIDCD NIH HHS/United States
- RF1 AG053303/AG/NIA NIH HHS/United States
- I01 BX003040/BX/BLRD VA/United States
- P30 AG012300/AG/NIA NIH HHS/United States
- RF1 AG051504/AG/NIA NIH HHS/United States
- P50 AG025688/AG/NIA NIH HHS/United States
- R01 AG041797/AG/NIA NIH HHS/United States
- R01 AG050603/AG/NIA NIH HHS/United States
- P30 AG013854/AG/NIA NIH HHS/United States
- R01 NS076837/NS/NINDS NIH HHS/United States
- R21 AG051839/AG/NIA NIH HHS/United States
- P30 AG010124/AG/NIA NIH HHS/United States
- P50 AG023501/AG/NIA NIH HHS/United States
- P50 AG005131/AG/NIA NIH HHS/United States
- R01 AG044546/AG/NIA NIH HHS/United States
- U24 AG021886/AG/NIA NIH HHS/United States
- MR/M008525/1/MRC_/Medical Research Council/United Kingdom
- P50 AG016574/AG/NIA NIH HHS/United States
- P01 AG017586/AG/NIA NIH HHS/United States
- R35 NS097261/NS/NINDS NIH HHS/United States
- P50 AG008702/AG/NIA NIH HHS/United States
- MC_UU_00024/1/MRC_/Medical Research Council/United Kingdom
- U24 NS095871/NS/NINDS NIH HHS/United States
- P01 AG003991/AG/NIA NIH HHS/United States
- P50 AG005681/AG/NIA NIH HHS/United States
- P01 AG026276/AG/NIA NIH HHS/United States
- P30 AG062429/AG/NIA NIH HHS/United States
- R01 AG051848/AG/NIA NIH HHS/United States
- P01 AG019724/AG/NIA NIH HHS/United States
- R01 NS080820/NS/NINDS NIH HHS/United States
- P50 NS072187/NS/NINDS NIH HHS/United States
- MR/J009482/1/MRC_/Medical Research Council/United Kingdom
- P50 AG005133/AG/NIA NIH HHS/United States
- MR/M023664/1/MRC_/Medical Research Council/United Kingdom
- RF1 AG044546/AG/NIA NIH HHS/United States
- P01 NS084974/NS/NINDS NIH HHS/United States
- U01 AG046139/AG/NIA NIH HHS/United States
- MR/L016397/1/MRC_/Medical Research Council/United Kingdom
- R01 AG037491/AG/NIA NIH HHS/United States
- U24 NS072026/NS/NINDS NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- U01 AG045390/AG/NIA NIH HHS/United States
- P30 AG019610/AG/NIA NIH HHS/United States
- U54 NS092089/NS/NINDS NIH HHS/United States
- U01 AG006786/AG/NIA NIH HHS/United States
- G0701441/MRC_/Medical Research Council/United Kingdom
- P30 NS055077/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

