Sequential and differential interaction of assembly factors during nitrogenase MoFe protein maturation
- PMID: 29724822
- PMCID: PMC6016461
- DOI: 10.1074/jbc.RA118.002994
Sequential and differential interaction of assembly factors during nitrogenase MoFe protein maturation
Abstract
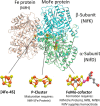
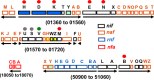
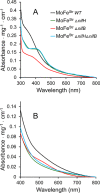
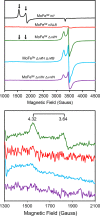
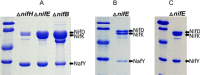
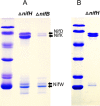
Nitrogenases reduce atmospheric nitrogen, yielding the basic inorganic molecule ammonia. The nitrogenase MoFe protein contains two cofactors, a [7Fe-9S-Mo-C-homocitrate] active-site species, designated FeMo-cofactor, and a [8Fe-7S] electron-transfer mediator called P-cluster. Both cofactors are essential for molybdenum-dependent nitrogenase catalysis in the nitrogen-fixing bacterium Azotobacter vinelandii We show here that three proteins, NafH, NifW, and NifZ, copurify with MoFe protein produced by an A. vinelandii strain deficient in both FeMo-cofactor formation and P-cluster maturation. In contrast, two different proteins, NifY and NafY, copurified with MoFe protein deficient only in FeMo-cofactor formation. We refer to proteins associated with immature MoFe protein in the following as "assembly factors." Copurifications of such assembly factors with MoFe protein produced in different genetic backgrounds revealed their sequential and differential interactions with MoFe protein during the maturation process. We found that these interactions occur in the order NafH, NifW, NifZ, and NafY/NifY. Interactions of NafH, NifW, and NifZ with immature forms of MoFe protein preceded completion of P-cluster maturation, whereas interaction of NafY/NifY preceded FeMo-cofactor insertion. Because each assembly factor could independently bind an immature form of MoFe protein, we propose that subpopulations of MoFe protein-assembly factor complexes represent MoFe protein captured at different stages of a sequential maturation process. This suggestion was supported by separate isolation of three such complexes, MoFe protein-NafY, MoFe protein-NifY, and MoFe protein-NifW. We conclude that factors involved in MoFe protein maturation sequentially bind and dissociate in a dynamic process involving several MoFe protein conformational states.
Keywords: FeMo-cofactor; MoFe protein; P-cluster; electron paramagnetic resonance (EPR); nitrogen fixation; nitrogenase; protein assembly; protein purification; reductase.
© 2018 Jimenez-Vicente et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

