Engineered 3-Ketosteroid 9α-Hydroxylases in Mycobacterium neoaurum: an Efficient Platform for Production of Steroid Drugs
- PMID: 29728384
- PMCID: PMC6029100
- DOI: 10.1128/AEM.02777-17
Engineered 3-Ketosteroid 9α-Hydroxylases in Mycobacterium neoaurum: an Efficient Platform for Production of Steroid Drugs
Abstract
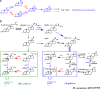
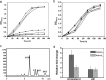
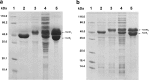
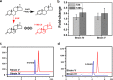
3-Ketosteroid 9α-hydroxylase (Ksh) consists of a terminal oxygenase (KshA) and a ferredoxin reductase and is indispensable in the cleavage of steroid nucleus in microorganisms. The activities of Kshs are crucial factors in determining the yield and distribution of products in the biotechnological transformation of sterols in industrial applications. In this study, two KshA homologues, KshA1N and KshA2N, were characterized and further engineered in a sterol-digesting strain, Mycobacterium neoaurum ATCC 25795, to construct androstenone-producing strains. kshA1N is a member of the gene cluster encoding sterol catabolism enzymes, and its transcription exhibited a 4.7-fold increase under cholesterol induction. Furthermore, null mutation of kshA1N led to the stable accumulation of androst-4-ene-3,17-dione (AD) and androst-1,4-diene-3,17-dione (ADD). We determined kshA2N to be a redundant form of kshA1N Through a combined modification of kshA1N, kshA2N, and other key genes involved in the metabolism of sterols, we constructed a high-yield ADD-producing strain that could produce 9.36 g liter-1 ADD from the transformation of 20 g liter-1 phytosterols in 168 h. Moreover, we improved a previously established 9α-hydroxy-AD-producing strain via the overexpression of a mutant KshA1N that had enhanced Ksh activity. Genetic engineering allowed the new strain to produce 11.7 g liter-1 9α-hydroxy-4-androstene-3,17-dione (9-OHAD) from the transformation of 20.0 g liter-1 phytosterol in 120 h.IMPORTANCE Steroidal drugs are widely used for anti-inflammation, anti-tumor action, endocrine regulation, and fertility management, among other uses. The two main starting materials for the industrial synthesis of steroid drugs are phytosterol and diosgenin. The phytosterol processing is carried out by microbial transformation, which is thought to be superior to the diosgenin processing by chemical conversions, given its simple and environmentally friendly process. However, diosgenin has long been used as the primary starting material instead of phytosterol. This is in response to challenges in developing efficient microbial strains for industrial phytosterol transformation, which stem from complex metabolic processes that feature many currently unclear details. In this study, we identified two oxygenase homologues of 3-ketosteroid-9α-hydroxylase, KshA1N and KshA2N, in M. neoaurum and demonstrated their crucial role in determining the yield and variety of products from phytosterol transformation. This work has practical value in developing industrial strains for phytosterol biotransformation.
Keywords: 3-ketosteroid-9α-hydroxylase; 9α-hydroxy-4-androstene-3,17-dione; 9α-hydroxy-4-androstene-3,17-dione (9-OHAD); Mn25795; androst-1,4-diene-3,17-dione; androst-1,4-diene-3,17-dione (ADD); mycobacterium; sterol; sterols.
Copyright © 2018 American Society for Microbiology.
Figures



Similar articles
-
Comparative analysis of genes encoding key steroid core oxidation enzymes in fast-growing Mycobacterium spp. strains.J Steroid Biochem Mol Biol. 2013 Nov;138:41-53. doi: 10.1016/j.jsbmb.2013.02.016. Epub 2013 Mar 6. J Steroid Biochem Mol Biol. 2013. PMID: 23474435
-
Efficient conversion of phytosterols into 4-androstene-3,17-dione and its C1,2-dehydrogenized and 9α-hydroxylated derivatives by engineered Mycobacteria.Microb Cell Fact. 2021 Aug 16;20(1):158. doi: 10.1186/s12934-021-01653-9. Microb Cell Fact. 2021. PMID: 34399754 Free PMC article.
-
Characterization and engineering of 3-ketosteroid-△1-dehydrogenase and 3-ketosteroid-9α-hydroxylase in Mycobacterium neoaurum ATCC 25795 to produce 9α-hydroxy-4-androstene-3,17-dione through the catabolism of sterols.Metab Eng. 2014 Jul;24:181-91. doi: 10.1016/j.ymben.2014.05.005. Epub 2014 May 14. Metab Eng. 2014. PMID: 24831710
-
3-Ketosteroid 9α-hydroxylase enzymes: Rieske non-heme monooxygenases essential for bacterial steroid degradation.Antonie Van Leeuwenhoek. 2014 Jul;106(1):157-72. doi: 10.1007/s10482-014-0188-2. Epub 2014 May 21. Antonie Van Leeuwenhoek. 2014. PMID: 24846050 Free PMC article. Review.
-
Biotransformation Enables Innovations Toward Green Synthesis of Steroidal Pharmaceuticals.ChemSusChem. 2022 May 6;15(9):e202102399. doi: 10.1002/cssc.202102399. Epub 2022 Feb 15. ChemSusChem. 2022. PMID: 35089653 Review.
Cited by
-
Whole-genome and enzymatic analyses of an androstenedione-producing Mycobacterium strain with residual phytosterol-degrading pathways.Microb Cell Fact. 2020 Oct 2;19(1):187. doi: 10.1186/s12934-020-01442-w. Microb Cell Fact. 2020. PMID: 33008397 Free PMC article.
-
Combined enhancement of the propionyl-CoA metabolic pathway for efficient androstenedione production in Mycolicibacterium neoaurum.Microb Cell Fact. 2022 Oct 20;21(1):218. doi: 10.1186/s12934-022-01942-x. Microb Cell Fact. 2022. PMID: 36266684 Free PMC article.
-
Rational development of mycobacteria cell factory for advancing the steroid biomanufacturing.World J Microbiol Biotechnol. 2022 Aug 17;38(11):191. doi: 10.1007/s11274-022-03369-3. World J Microbiol Biotechnol. 2022. PMID: 35974205 Free PMC article. Review.
-
Production of 21-hydroxy-20-methyl-pregna-1,4-dien-3-one by modifying multiple genes in Mycolicibacterium.Appl Microbiol Biotechnol. 2023 Mar;107(5-6):1563-1574. doi: 10.1007/s00253-023-12399-2. Epub 2023 Feb 2. Appl Microbiol Biotechnol. 2023. PMID: 36729227
-
Production of 11α-hydroxysteroids from sterols in a single fermentation step by Mycolicibacterium smegmatis.Microb Biotechnol. 2021 Nov;14(6):2514-2524. doi: 10.1111/1751-7915.13735. Epub 2021 Mar 4. Microb Biotechnol. 2021. PMID: 33660943 Free PMC article.
References
-
- Yao K, Xu LQ, Wang FQ, Wei DZ. 2014. Characterization and engineering of 3-ketosteroid-Δ1-dehydrogenase and 3-ketosteroid-9α-hydroxylase in Mycobacterium neoaurum ATCC 25795 to produce 9α-hydroxy-4-androstene-3, 17-dione through the catabolism of sterols. Metab Eng 24:181–191. doi:10.1016/j.ymben.2014.05.005. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

