A bipartite periplasmic receptor-diguanylate cyclase pair (XAC2383-XAC2382) in the bacterium Xanthomonas citri
- PMID: 29728456
- PMCID: PMC6036220
- DOI: 10.1074/jbc.RA118.003475
A bipartite periplasmic receptor-diguanylate cyclase pair (XAC2383-XAC2382) in the bacterium Xanthomonas citri
Abstract
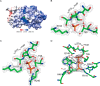
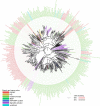
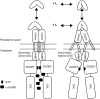
The second messenger cyclic diguanylate monophosphate (c-di-GMP) is a central regulator of bacterial lifestyle, controlling several behaviors, including the switch between sessile and motile states. The c-di-GMP levels are controlled by the interplay between diguanylate cyclases (DGCs) and phosphodiesterases, which synthesize and hydrolyze this second messenger, respectively. These enzymes often contain additional domains that regulate activity via binding of small molecules, covalent modification, or protein-protein interactions. A major challenge remains to understand how DGC activity is regulated by these additional domains or interaction partners in specific signaling pathways. Here, we identified a pair of co-transcribed genes (xac2382 and xac2383) in the phytopathogenic, Gram-negative bacterium Xanthomonas citri subsp. citri (Xac), whose mutations resulted in opposing motility phenotypes. We show that the periplasmic cache domain of XAC2382, a membrane-associated DGC, interacts with XAC2383, a periplasmic binding protein, and we provide evidence that this interaction regulates XAC2382 DGC activity. Moreover, we solved the crystal structure of XAC2383 with different ligands, indicating a preference for negatively charged phosphate-containing compounds. We propose that XAC2383 acts as a periplasmic sensor that, upon binding its ligand, inhibits the DGC activity of XAC2382. Of note, we also found that this previously uncharacterized signal transduction system is present in several other bacterial phyla, including Gram-positive bacteria. Phylogenetic analysis of homologs of the XAC2382-XAC2383 pair supports several independent origins that created new combinations of XAC2382 homologs with a conserved periplasmic cache domain with different cytoplasmic output module architectures.
Keywords: Xanthomonas; bacterial signal transduction; cell motility; cyclic di-GMP (c-di-GMP); phytopathogen; plant pathogen; receptor; structural biology.
© 2018 Teixeira et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures

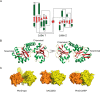



References
-
- Whitney J. C., Colvin K. M., Marmont L. S., Robinson H., Parsek M. R., and Howell P. L. (2012) Structure of the cytoplasmic region of PelD, a degenerate diguanylate cyclase receptor that regulates exopolysaccharide production in Pseudomonas aeruginosa. J. Biol. Chem. 287, 23582–23593 10.1074/jbc.M112.375378 - DOI - PMC - PubMed
-
- Kuchma S. L., Brothers K. M., Merritt J. H., Liberati N. T., Ausubel F. M., and O'Toole G. A. (2007) BifA, a cyclic-di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 189, 8165–8178 10.1128/JB.00586-07 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

