Promoter of lncRNA Gene PVT1 Is a Tumor-Suppressor DNA Boundary Element
- PMID: 29731168
- PMCID: PMC5984165
- DOI: 10.1016/j.cell.2018.03.068
Promoter of lncRNA Gene PVT1 Is a Tumor-Suppressor DNA Boundary Element
Abstract
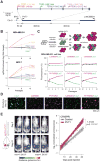
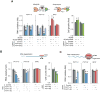
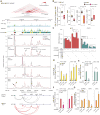
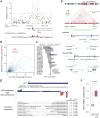
Noncoding mutations in cancer genomes are frequent but challenging to interpret. PVT1 encodes an oncogenic lncRNA, but recurrent translocations and deletions in human cancers suggest alternative mechanisms. Here, we show that the PVT1 promoter has a tumor-suppressor function that is independent of PVT1 lncRNA. CRISPR interference of PVT1 promoter enhances breast cancer cell competition and growth in vivo. The promoters of the PVT1 and the MYC oncogenes, located 55 kb apart on chromosome 8q24, compete for engagement with four intragenic enhancers in the PVT1 locus, thereby allowing the PVT1 promoter to regulate pause release of MYC transcription. PVT1 undergoes developmentally regulated monoallelic expression, and the PVT1 promoter inhibits MYC expression only from the same chromosome via promoter competition. Cancer genome sequencing identifies recurrent mutations encompassing the human PVT1 promoter, and genome editing verified that PVT1 promoter mutation promotes cancer cell growth. These results highlight regulatory sequences of lncRNA genes as potential disease-associated DNA elements.
Keywords: CRISPRi; MYC; PVT1; enhancer; lncRNA; promoter; topological domains; transcriptional regulation; tumor suppressor.
Copyright © 2018 Elsevier Inc. All rights reserved.
Figures



Comment in
-
A "Counter-Enhancer" in Tumor Suppression.Cell. 2018 May 31;173(6):1318-1319. doi: 10.1016/j.cell.2018.05.023. Epub 2018 May 31. Cell. 2018. PMID: 29856950
-
Competing for enhancers: PVT1 fine-tunes MYC expression.Cell Res. 2018 Aug;28(8):785-786. doi: 10.1038/s41422-018-0064-0. Cell Res. 2018. PMID: 29980733 Free PMC article. No abstract available.
References
-
- Blancafort P, Magnenat L, Barbas CF., 3rd Scanning the human genome with combinatorial transcription factor libraries. Nat Biotechnol. 2003;21:269–274. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

