An integrative model for alternative polyadenylation, IntMAP, delineates mTOR-modulated endoplasmic reticulum stress response
- PMID: 29733382
- PMCID: PMC6158760
- DOI: 10.1093/nar/gky340
An integrative model for alternative polyadenylation, IntMAP, delineates mTOR-modulated endoplasmic reticulum stress response
Abstract
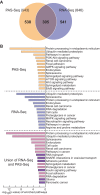
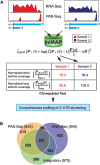
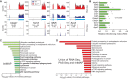
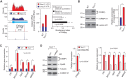
3'-untranslated regions (UTRs) can vary through the use of alternative polyadenylation sites during pre-mRNA processing. Multiple publically available pipelines combining high profiling technologies and bioinformatics tools have been developed to catalog changes in 3'-UTR lengths. In our recent RNA-seq experiments using cells with hyper-activated mammalian target of rapamycin (mTOR), we found that cellular mTOR activation leads to transcriptome-wide alternative polyadenylation (APA), resulting in the activation of multiple cellular pathways. Here, we developed a novel bioinformatics algorithm, IntMAP, which integrates RNA-Seq and PolyA Site (PAS)-Seq data for a comprehensive characterization of APA events. By applying IntMAP to the datasets from cells with hyper-activated mTOR, we identified novel APA events that could otherwise not be identified by either profiling method alone. Several transcription factors including Cebpg (CCAAT/enhancer binding protein gamma) were among the newly discovered APA transcripts, indicating that diverse transcriptional networks may be regulated by mTOR-coordinated APA. The prevention of APA in Cebpg using the CRISPR/cas9-mediated genome editing tool showed that mTOR-driven 3'-UTR shortening in Cebpg is critical in protecting cells from endoplasmic reticulum (ER) stress. Taken together, we present IntMAP as a new bioinformatics algorithm for APA analysis by which we expand our understanding of the physiological role of mTOR-coordinated APA events to ER stress response. IntMAP toolbox is available at http://compbio.cs.umn.edu/IntMAP/.
Figures

References
-
- Manley J.L. Polyadenylation of mRNA precursors. BBA - Gene Struct. Expr. 1988; 950:1–12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

