Cryo-EM shows stages of initial codon selection on the ribosome by aa-tRNA in ternary complex with GTP and the GTPase-deficient EF-TuH84A
- PMID: 29733411
- PMCID: PMC6009598
- DOI: 10.1093/nar/gky346
Cryo-EM shows stages of initial codon selection on the ribosome by aa-tRNA in ternary complex with GTP and the GTPase-deficient EF-TuH84A
Abstract
The GTPase EF-Tu in ternary complex with GTP and aminoacyl-tRNA (aa-tRNA) promotes rapid and accurate delivery of cognate aa-tRNAs to the ribosomal A site. Here we used cryo-EM to study the molecular origins of the accuracy of ribosome-aided recognition of a cognate ternary complex and the accuracy-amplifying role of the monitoring bases A1492, A1493 and G530 of the 16S rRNA. We used the GTPase-deficient EF-Tu variant H84A with native GTP, rather than non-cleavable GTP analogues, to trap a near-cognate ternary complex in high-resolution ribosomal complexes of varying codon-recognition accuracy. We found that ribosome complexes trapped by GTPase-deficicent ternary complex due to the presence of EF-TuH84A or non-cleavable GTP analogues have very similar structures. We further discuss speed and accuracy of initial aa-tRNA selection in terms of conformational changes of aa-tRNA and stepwise activation of the monitoring bases at the decoding center of the ribosome.
Figures
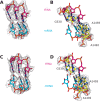




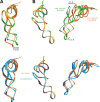

Similar articles
-
Structural dynamics of translation elongation factor Tu during aa-tRNA delivery to the ribosome.Nucleic Acids Res. 2018 Sep 19;46(16):8651-8661. doi: 10.1093/nar/gky651. Nucleic Acids Res. 2018. PMID: 30107527 Free PMC article.
-
Distinct functional classes of ram mutations in 16S rRNA.RNA. 2014 Apr;20(4):496-504. doi: 10.1261/rna.043331.113. Epub 2014 Feb 26. RNA. 2014. PMID: 24572811 Free PMC article.
-
Elongation factor Tu, a GTPase triggered by codon recognition on the ribosome: mechanism and GTP consumption.Biochem Cell Biol. 1995 Nov-Dec;73(11-12):1221-7. doi: 10.1139/o95-132. Biochem Cell Biol. 1995. PMID: 8722040 Review.
-
Steric complementarity in the decoding center is important for tRNA selection by the ribosome.J Mol Biol. 2013 Oct 23;425(20):3778-89. doi: 10.1016/j.jmb.2013.02.038. Epub 2013 Mar 27. J Mol Biol. 2013. PMID: 23542008 Free PMC article.
-
Recognition and selection of tRNA in translation.FEBS Lett. 2005 Feb 7;579(4):938-42. doi: 10.1016/j.febslet.2004.11.048. FEBS Lett. 2005. PMID: 15680978 Review.
Cited by
-
Transient disome complex formation in native polysomes during ongoing protein synthesis captured by cryo-EM.Nat Commun. 2024 Feb 26;15(1):1756. doi: 10.1038/s41467-024-46092-3. Nat Commun. 2024. PMID: 38409277 Free PMC article.
-
The Structural Dynamics of Translation.Annu Rev Biochem. 2022 Jun 21;91:245-267. doi: 10.1146/annurev-biochem-071921-122857. Epub 2022 Mar 14. Annu Rev Biochem. 2022. PMID: 35287473 Free PMC article. Review.
-
Genetic code degeneracy is established by the decoding center of the ribosome.Nucleic Acids Res. 2022 Apr 22;50(7):4113-4126. doi: 10.1093/nar/gkac171. Nucleic Acids Res. 2022. PMID: 35325219 Free PMC article.
-
tRNA Dissociation from EF-Tu after GTP Hydrolysis: Primary Steps and Antibiotic Inhibition.Biophys J. 2020 Jan 7;118(1):151-161. doi: 10.1016/j.bpj.2019.10.028. Epub 2019 Oct 28. Biophys J. 2020. PMID: 31711607 Free PMC article.
-
Advances in domain and subunit localization technology for electron microscopy.Biophys Rev. 2019 Apr;11(2):149-155. doi: 10.1007/s12551-019-00513-6. Epub 2019 Mar 5. Biophys Rev. 2019. PMID: 30834502 Free PMC article. Review.
References
-
- Voorhees R.M., Ramakrishnan V.. Structural basis of the translational elongation cycle. Annu. Rev. Biochem. 2013; 82:203–236. - PubMed
-
- Kavaliauskas D., Nissen P., Knudsen C.R.. The busiest of all ribosomal assistants: elongation factor Tu. Biochemistry. 2012; 51:2642–2651. - PubMed
-
- Ogle J.M., Brodersen D.E., Clemons W.M. Jr, Tarry M.J., Carter A.P., Ramakrishnan V.. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001; 292:897–902. - PubMed
-
- Ogle J.M., Murphy F.V., Tarry M.J., Ramakrishnan V.. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002; 111:721–732. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

