Linked genetic variation and not genome structure causes widespread differential expression associated with chromosomal inversions
- PMID: 29735663
- PMCID: PMC6003460
- DOI: 10.1073/pnas.1721275115
Linked genetic variation and not genome structure causes widespread differential expression associated with chromosomal inversions
Abstract
Chromosomal inversions are widely thought to be favored by natural selection because they suppress recombination between alleles that have higher fitness on the same genetic background or in similar environments. Nonetheless, few selected alleles have been characterized at the molecular level. Gene expression profiling provides a powerful way to identify functionally important variation associated with inversions and suggests candidate phenotypes. However, altered genome structure itself might also impact gene expression by influencing expression profiles of the genes proximal to inversion breakpoint regions or by modifying expression patterns genome-wide due to rearranging large regulatory domains. In natural inversions, genetic differentiation and genome structure are inextricably linked. Here, we characterize differential expression patterns associated with two chromosomal inversions found in natural Drosophila melanogaster populations. To isolate the impacts of genome structure, we engineered synthetic chromosomal inversions on controlled genetic backgrounds with breakpoints that closely match each natural inversion. We find that synthetic inversions have negligible effects on gene expression. Nonetheless, natural inversions have broad-reaching regulatory impacts in cis and trans Furthermore, we find that differentially expressed genes associated with both natural inversions are enriched for loci associated with immune response to bacterial pathogens. Our results support the idea that inversions in D. melanogaster experience natural selection to maintain associations between functionally related alleles to produce complex phenotypic outcomes.
Keywords: chromosomal inversions; differential expression; genome structure.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

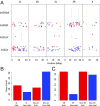
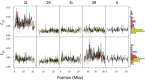
References
-
- Charlesworth D, Charlesworth B, Marais G. Steps in the evolution of heteromorphic sex chromosomes. Heredity (Edinb) 2005;95:118–128. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

