Universality of clone dynamics during tissue development
- PMID: 29736183
- PMCID: PMC5935228
- DOI: 10.1038/s41567-018-0055-6
Universality of clone dynamics during tissue development
Abstract
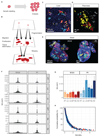
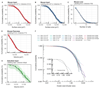
The emergence of complex organs is driven by the coordinated proliferation, migration and differentiation of precursor cells. The fate behaviour of these cells is reflected in the time evolution their progeny, termed clones, which serve as a key experimental observable. In adult tissues, where cell dynamics is constrained by the condition of homeostasis, clonal tracing studies based on transgenic animal models have advanced our understanding of cell fate behaviour and its dysregulation in disease (1, 2). But what can be learned from clonal dynamics in development, where the spatial cohesiveness of clones is impaired by tissue deformations during tissue growth? Drawing on the results of clonal tracing studies, we show that, despite the complexity of organ development, clonal dynamics may converge to a critical state characterized by universal scaling behaviour of clone sizes. By mapping clonal dynamics onto a generalization of the classical theory of aerosols, we elucidate the origin and range of scaling behaviours and show how the identification of universal scaling dependences may allow lineage-specific information to be distilled from experiments. Our study shows the emergence of core concepts of statistical physics in an unexpected context, identifying cellular systems as a laboratory to study non-equilibrium statistical physics.
Conflict of interest statement
Competing financial interests: The authors declare no competing financial interests.
Figures

References
-
- Kretzschmar K, Watt FM. Lineage tracing. Cell. 2012;148:33–45. - PubMed
-
- Blanpain C, Simons BD. Unravelling stem cell dynamics by lineage tracing. Nat Rev Mol Cell Biol. 2013;14:489–502. - PubMed
-
- Klein AM, Simons BD. Universal patterns of stem cell fate in cycling adult tissues. Development. 2011;138:3103–11. - PubMed
-
- Rulands S, Simons BD. Tracing cellular dynamics in tissue development, maintenance and disease. Curr Opin Cell Biol. 2016;43:38–45. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
