LncRNA MALAT1 promotes tumor growth and metastasis by targeting miR-124/foxq1 in bladder transitional cell carcinoma (BTCC)
- PMID: 29736319
- PMCID: PMC5934564
LncRNA MALAT1 promotes tumor growth and metastasis by targeting miR-124/foxq1 in bladder transitional cell carcinoma (BTCC)
Abstract
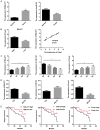
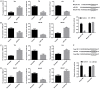
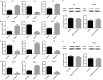
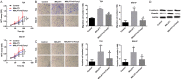
Mounting evidence shows that the long non-coding RNA MALAT1 plays a pivotal role in tumorigenesis and metastasis, but the functional significance of MALAT1 in bladder transitional cell carcinoma (BTCC) remains unclear. MALAT1 expression was measured in 56 BTCC patients and 2 BTCC cell lines by real-time PCR. The effects of MALAT1 on BTCC cells were investigated by over-expression approaches in vitro and in vivo. Insights of the mechanism of competitive endogenous RNAs (ceRNAs) were validated through bioinformatic analysis and luciferase assay. MALAT1 up-regulation positively correlated with advanced clinical pathological stage and shorter survival of BTCC patients. Furthermore, MALAT1 over-expression promoted proliferation, migration and invasion of BTCC cells in vitro and in vivo. Particularly, MALAT1 may function as a ceRNA to sponge miR-124, thus modulating the derepression of foxq1, miR-124 target gene, in post-transcriptional levels. The positive MALAT1/foxq1 interaction was confirmed by bivariate correlation analysis, and this positive correlation was of great significance in BTCC tumor growth and metastasis, also accompanied by EMT changes. Overall, this ceRNA regulatory network concerning MALAT1 and the positive MALAT1/foxq1 correlation benefit a better understanding of BTCC pathogenesis and promote the feasibility of lncRNA-directed therapy against this disease.
Keywords: Bladder transitional cell carcinoma (BTCC); MALAT1; competing endogenous RNA(ceRNA); foxq1; proliferation and invasion.
Conflict of interest statement
None.
Figures
Similar articles
-
Short hairpin RNA targeting FOXQ1 inhibits invasion and metastasis via the reversal of epithelial-mesenchymal transition in bladder cancer.Int J Oncol. 2013 Apr;42(4):1271-8. doi: 10.3892/ijo.2013.1807. Epub 2013 Feb 5. Int J Oncol. 2013. PMID: 23403865
-
Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer.Mol Cancer. 2014 Apr 28;13:92. doi: 10.1186/1476-4598-13-92. Mol Cancer. 2014. PMID: 24775712 Free PMC article.
-
Long noncoding RNA GAS5 inhibits malignant proliferation and chemotherapy resistance to doxorubicin in bladder transitional cell carcinoma.Cancer Chemother Pharmacol. 2017 Jan;79(1):49-55. doi: 10.1007/s00280-016-3194-4. Epub 2016 Nov 22. Cancer Chemother Pharmacol. 2017. Retraction in: Cancer Chemother Pharmacol. 2025 Feb 3;95(1):30. doi: 10.1007/s00280-025-04758-9. PMID: 27878359 Retracted.
-
MicroRNA-497 inhibits the proliferation, migration and invasion of human bladder transitional cell carcinoma cells by targeting E2F3.Oncol Rep. 2016 Sep;36(3):1293-300. doi: 10.3892/or.2016.4923. Epub 2016 Jul 8. Oncol Rep. 2016. PMID: 27430325
-
Beyond the Genome: Deciphering the Role of MALAT1 in Breast Cancer Progression.Curr Genomics. 2024;25(5):343-357. doi: 10.2174/0113892029305656240503045154. Epub 2024 May 22. Curr Genomics. 2024. PMID: 39323624 Free PMC article. Review.
Cited by
-
Bone Marrow Mesenchymal Stem Cells-Derived Extracellular Vesicles Promote Proliferation, Invasion and Migration of Osteosarcoma Cells via the lncRNA MALAT1/miR-143/NRSN2/Wnt/β-Catenin Axis.Onco Targets Ther. 2021 Feb 2;14:737-749. doi: 10.2147/OTT.S283459. eCollection 2021. Onco Targets Ther. 2021. PMID: 33564242 Free PMC article.
-
The functions of microRNA-124 on bladder cancer.Onco Targets Ther. 2019 May 7;12:3429-3439. doi: 10.2147/OTT.S193661. eCollection 2019. Onco Targets Ther. 2019. PMID: 31190856 Free PMC article.
-
Long non-coding RNAs: regulators of autophagy and potential biomarkers in therapy resistance and urological cancers.Front Pharmacol. 2024 Oct 24;15:1442227. doi: 10.3389/fphar.2024.1442227. eCollection 2024. Front Pharmacol. 2024. PMID: 39512820 Free PMC article. Review.
-
The mechanism of lncRNA-CRNDE in regulating tumour-associated macrophage M2 polarization and promoting tumour angiogenesis.J Cell Mol Med. 2021 May;25(9):4235-4247. doi: 10.1111/jcmm.16477. Epub 2021 Mar 20. J Cell Mol Med. 2021. PMID: 33742511 Free PMC article.
-
Function of microRNA‑124 in the pathogenesis of cancer (Review).Int J Oncol. 2024 Jan;64(1):6. doi: 10.3892/ijo.2023.5594. Epub 2023 Dec 1. Int J Oncol. 2024. PMID: 38038165 Free PMC article. Review.
References
-
- Murta-Nascimento C, Schmitz-Dräger BJ, Zeegers MP, Steineck G, Kogevinas M, Real FX, Malats N. Epidemiology of urinary bladder cancer: from tumor development to patient’s death. World J Urol. 2007;25:285–295. - PubMed
-
- Kim WJ, Quan C. Genetic and epigenetic aspects of bladder cancer. J Cell Biochem. 2005;95:24–33. - PubMed
-
- Wang HT, Chang JW. Molecular pathology of low malignant bladder transitional cell carcinoma: a current perspective. Histol Histopathol. 2005;20:147–153. - PubMed
-
- Nordentoft I, Lamy P, Birkenkamp-Demtröder K, Shumansky K, Vang S, Hornshøj H, Juul M, Villesen P, Hedegaard J, Roth A. Mutational context and diverse clonal development in early and late bladder cancer. Cell Rep. 2014;7:1649–1663. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
