Hidden MHC genetic diversity in the Iberian ibex (Capra pyrenaica)
- PMID: 29739323
- PMCID: PMC5941765
- DOI: 10.1186/s12863-018-0616-9
Hidden MHC genetic diversity in the Iberian ibex (Capra pyrenaica)
Abstract
Background: Defining hidden genetic diversity within species is of great significance when attempting to maintain the evolutionary potential of natural populations and conduct appropriate management. Our hypothesis is that isolated (and eventually small) wild animal populations hide unexpected genetic diversity due to their maintenance of ancient polymorphisms or introgressions.
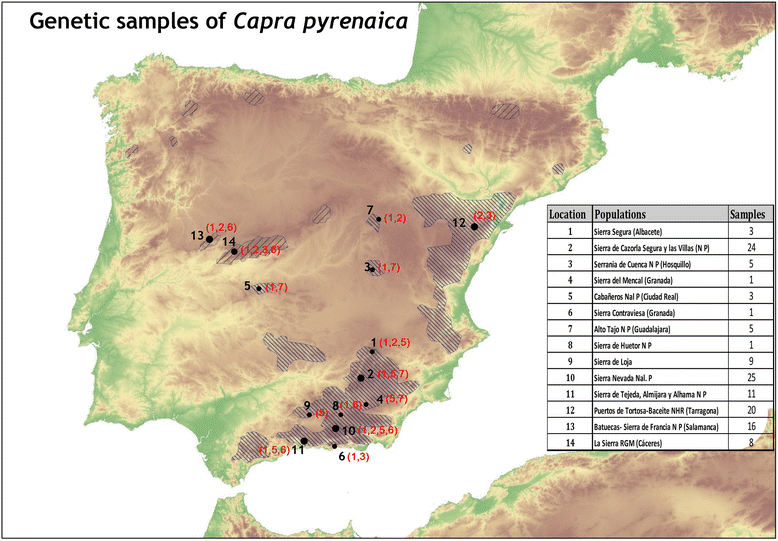
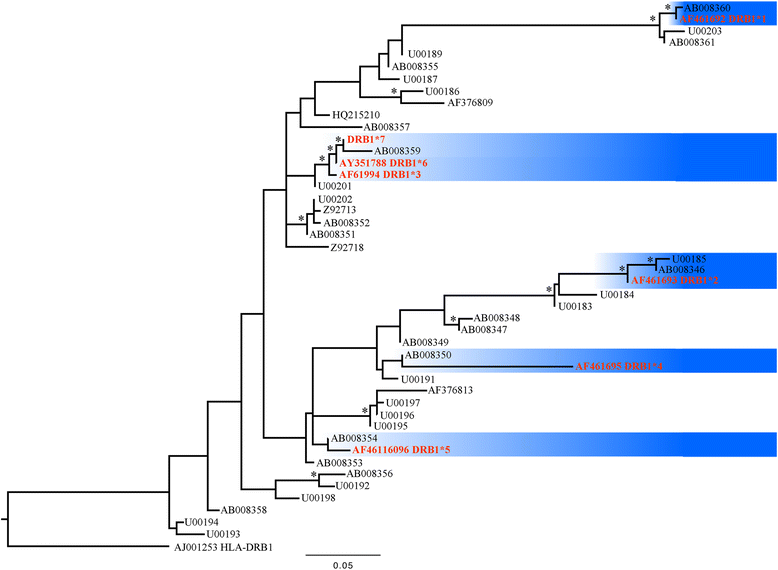
Results: We tested this hypothesis using the Iberian ibex (Capra pyrenaica) as an example. Previous studies based on large sample sizes taken from its principal populations have revealed that the Iberian ibex has a remarkably small MHC DRB1 diversity (only six remnant alleles) as a result of recent population bottlenecks and a marked demographic decline that has led to the extinction of two recognized subspecies. Extending on the geographic range to include non-studied isolated Iberian ibex populations, we sequenced a new MHC DRB1 in what seemed three small isolated populations in Southern Spain (n = 132). The findings indicate a higher genetic diversity than previously reported in this important gene. The newly discovered allele, MHC DRB1*7, is identical to one reported in the domestic goat C. aegagrus hircus. Whether or not this is the result of ancient polymorphisms maintained by balancing selection or, alternatively, introgressions from domestic goats through hybridization needs to be clarified in future studies. However, hybridization between Iberian ibex and domestic goats has been reported in Spain and the fact that the newly discovered allele is only present in one of the small isolated populations and not in the others suggests introgression. The new discovered allele is not expected to increase fitness in C. pyrenaica since it generates the same protein as the existing MHC DRB1*6. Analysis of a microsatellite locus (OLADRB1) near the new MHC DRB1*7 gene reveals a linkage disequilibrium between these two loci. The allele OLADRB1, 187 bp in length, was unambiguously linked to the MHC DRB1*7 allele. This enabled us to perform a DRB-STR matching method for the recently discovered MHC allele.
Conclusions: This finding is critical for the conservation of the Iberian ibex since it directly affects the identification of the units of this species that should be managed and conserved separately (Evolutionarily Significant Units).
Keywords: Capra aegagrus hircus; Capra pyrenaica hispanica; Capra pyrenaica victoriae; DRB-STR method; Linkage disequilibrium; MHC DRB1; Major histocompatibility complex (MHC); OLADRB1; Segura and las Villas Natural Park; Sierras de Cazorla; Spain.
Conflict of interest statement
Ethics approval
The samples consisted of tissue (small biopsy from the ears) obtained from deceased legally hunted animals, or from animals culled by park rangers as part of wildlife management plans aimed especially at controlling ungulate density and preventing outbreaks of diseases. Thus, no animal ethical permits were necessary since no live ibex were handled during this study. This study was approved by the Spanish Ministry of Agriculture, Fishery and Environment of the Andalusian government (Junta de Andalucía, Spain). Sampling procedures were issued as part of the application for permits for the fieldwork, which did not affect any endangered or protected species.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- Carvalho DC, Denise Oliveira AA, Behegaray LB, Torres RA. Hidden genetic diversity and distinct evolutionarily significant units in an commercially important Neotropical apex predator, the catfish Pseudoplatystoma corruscans. Conserv Genet. 2012;13:1671–1675. doi: 10.1007/s10592-012-0402-6. - DOI
-
- Frankham R. Challenges and opportunities of genetic approaches to biological conservation. Biol Conserv. 2010;143:1919–1927. doi: 10.1016/j.biocon.2010.05.011. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

