Recurrent hotspot mutations in HRAS Q61 and PI3K-AKT pathway genes as drivers of breast adenomyoepitheliomas
- PMID: 29739933
- PMCID: PMC5940840
- DOI: 10.1038/s41467-018-04128-5
Recurrent hotspot mutations in HRAS Q61 and PI3K-AKT pathway genes as drivers of breast adenomyoepitheliomas
Abstract
Adenomyoepithelioma of the breast is a rare tumor characterized by epithelial-myoepithelial differentiation, whose genetic underpinning is largely unknown. Here we show through whole-exome and targeted massively parallel sequencing analysis that whilst estrogen receptor (ER)-positive adenomyoepitheliomas display PIK3CA or AKT1 activating mutations, ER-negative adenomyoepitheliomas harbor highly recurrent codon Q61 HRAS hotspot mutations, which co-occur with PIK3CA or PIK3R1 mutations. In two- and three-dimensional cell culture models, forced expression of HRASQ61R in non-malignant ER-negative breast epithelial cells with or without a PIK3CAH1047R somatic knock-in results in transformation and the acquisition of the cardinal features of adenomyoepitheliomas, including the expression of myoepithelial markers, a reduction in E-cadherin expression, and an increase in AKT signaling. Our results demonstrate that adenomyoepitheliomas are genetically heterogeneous, and qualify mutations in HRAS, a gene whose mutations are vanishingly rare in common-type breast cancers, as likely drivers of ER-negative adenomyoepitheliomas.
Conflict of interest statement
The authors declare no competing interests.
Figures
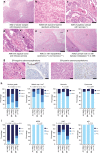



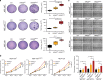
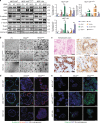
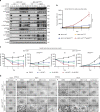
References
-
- Lakhani SR, Ellis IO, Schnitt SJ, Tan PH, van de Vijver MJ. WHO Classification of Tumours of the Breast. Lyon: IARC; 2012.
-
- Nadelman CM, Leslie KO, Fishbein MC. “Benign,” metastasizing adenomyoepithelioma of the breast: a report of 2 cases. Arch. Pathol. Lab. Med. 2006;130:1349–1353. - PubMed
-
- Moritz AW, Wiedenhoefer JF, Profit AP, Jagirdar J. Breast adenomyoepithelioma and adenomyoepithelioma with carcinoma (malignant adenomyoepithelioma) with associated breast malignancies: a case series emphasizing histologic, radiologic, and clinical correlation. Breast. 2016;29:132–139. doi: 10.1016/j.breast.2016.07.018. - DOI - PubMed
-
- Ng CK, Schultheis AM, Bidard FC, Weigelt B, Reis-Filho JS. Breast cancer genomics from microarrays to massively parallel sequencing: paradigms and new insights. J. Natl. Cancer Inst. 2015;107:djv015. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

