Aromatic Rings Commonly Used in Medicinal Chemistry: Force Fields Comparison and Interactions With Water Toward the Design of New Chemical Entities
- PMID: 29740321
- PMCID: PMC5928326
- DOI: 10.3389/fphar.2018.00395
Aromatic Rings Commonly Used in Medicinal Chemistry: Force Fields Comparison and Interactions With Water Toward the Design of New Chemical Entities
Abstract
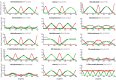
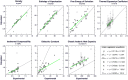
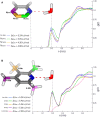
The identification of lead compounds usually includes a step of chemical diversity generation. Its rationale may be supported by both qualitative (SAR) and quantitative (QSAR) approaches, offering models of the putative ligand-receptor interactions. In both scenarios, our understanding of which interactions functional groups can perform is mostly based on their chemical nature (such as electronegativity, volume, melting point, lipophilicity etc.) instead of their dynamics in aqueous, biological solutions (solvent accessibility, lifetime of hydrogen bonds, solvent structure etc.). As a consequence, it is challenging to predict from 2D structures which functional groups will be able to perform interactions with the target receptor, at which intensity and relative abundance in the biological environment, all of which will contribute to ligand potency and intrinsic activity. With this in mind, the aim of this work is to assess properties of aromatic rings, commonly used for drug design, in aqueous solution through molecular dynamics simulations in order to characterize their chemical features and infer their impact in complexation dynamics. For this, common aromatic and heteroaromatic rings were selected and received new atomic charge set based on the direction and module of the dipole moment from MP2/6-31G* calculations, while other topological terms were taken from GROMOS53A6 force field. Afterwards, liquid physicochemical properties were simulated for a calibration set composed by nearly 40 molecules and compared to their respective experimental data, in order to validate each topology. Based on the reliance of the employed strategy, we expanded the dataset to more than 100 aromatic rings. Properties in aqueous solution such as solvent accessible surface area, H-bonds availability, H-bonds residence time, and water structure around heteroatoms were calculated for each ring, creating a database of potential interactions, shedding light on features of drugs in biological solutions, on the structural basis for bioisosterism and on the enthalpic/entropic costs for ligand-receptor complexation dynamics.
Keywords: GROMOS; aromatic rings; drug design; functional groups; interactions.
Figures
References
-
- Abraham M. J., Murtola T., Schulz R., Páll S., Smith J. C., Hess B., et al. (2015). Gromacs: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2, 19–25. 10.1016/j.softx.2015.06.001 - DOI
-
- Aqvist J., Medina C., Samuelsson J. E. (1994). A new method for predicting binding affinity in computer-aided drug design. Protein Eng. 7, 385–391. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

