Overview of Virus Metagenomic Classification Methods and Their Biological Applications
- PMID: 29740407
- PMCID: PMC5924777
- DOI: 10.3389/fmicb.2018.00749
Overview of Virus Metagenomic Classification Methods and Their Biological Applications
Abstract
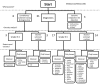
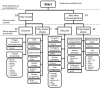
Metagenomics poses opportunities for clinical and public health virology applications by offering a way to assess complete taxonomic composition of a clinical sample in an unbiased way. However, the techniques required are complicated and analysis standards have yet to develop. This, together with the wealth of different tools and workflows that have been proposed, poses a barrier for new users. We evaluated 49 published computational classification workflows for virus metagenomics in a literature review. To this end, we described the methods of existing workflows by breaking them up into five general steps and assessed their ease-of-use and validation experiments. Performance scores of previous benchmarks were summarized and correlations between methods and performance were investigated. We indicate the potential suitability of the different workflows for (1) time-constrained diagnostics, (2) surveillance and outbreak source tracing, (3) detection of remote homologies (discovery), and (4) biodiversity studies. We provide two decision trees for virologists to help select a workflow for medical or biodiversity studies, as well as directions for future developments in clinical viral metagenomics.
Keywords: decision tree; pipeline; software; standardization; use case; viral metagenomics.
Figures




References
-
- Alves J. M., de Oliveira A. L., Sandberg T. O., Moreno-Gallego J. L., de Toledo M. A., de Moura E. M., et al. . (2016). GenSeed-HMM: a tool for progressive assembly using profile HMMs as seeds and its application in alpavirinae viral discovery from metagenomic data. Front. Microbiol. 7:269. 10.3389/fmicb.2016.00269 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

