The Membrane-Bound C Subunit of Reductive Dehalogenases: Topology Analysis and Reconstitution of the FMN-Binding Domain of PceC
- PMID: 29740408
- PMCID: PMC5928378
- DOI: 10.3389/fmicb.2018.00755
The Membrane-Bound C Subunit of Reductive Dehalogenases: Topology Analysis and Reconstitution of the FMN-Binding Domain of PceC
Abstract
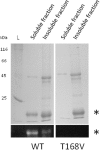
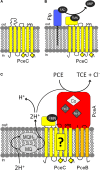
Organohalide respiration (OHR) is the energy metabolism of anaerobic bacteria able to use halogenated organic compounds as terminal electron acceptors. While the terminal enzymes in OHR, so-called reductive dehalogenases, are well-characterized, the identity of proteins potentially involved in electron transfer to the terminal enzymes remains elusive. Among the accessory genes identified in OHR gene clusters, the C subunit (rdhC) could well code for the missing redox protein between the quinol pool and the reductive dehalogenase, although it was initially proposed to act as transcriptional regulator. RdhC sequences are characterized by the presence of multiple transmembrane segments, a flavin mononucleotide (FMN) binding motif and two conserved CX3CP motifs. Based on these features, we propose a curated selection of RdhC proteins identified in general sequence databases. Beside the Firmicutes from which RdhC sequences were initially identified, the identified sequences belong to three additional phyla, the Chloroflexi, the Proteobacteria, and the Bacteriodetes. The diversity of RdhC sequences mostly respects the phylogenetic distribution, suggesting that rdhC genes emerged relatively early in the evolution of the OHR metabolism. PceC, the C subunit of the tetrachloroethene (PCE) reductive dehalogenase is encoded by the conserved pceABCT gene cluster identified in Dehalobacter restrictus PER-K23 and in several strains of Desulfitobacterium hafniense. Surfaceome analysis of D. restrictus cells confirmed the predicted topology of the FMN-binding domain (FBD) of PceC that is the exocytoplasmic face of the membrane. Starting from inclusion bodies of a recombinant FBD protein, strategies for successful assembly of the FMN cofactor and refolding were achieved with the use of the flavin-trafficking protein from D. hafniense TCE1. Mass spectrometry analysis and site-directed mutagenesis of rFBD revealed that threonine-168 of PceC is binding FMN covalently. Our results suggest that PceC, and more generally RdhC proteins, may play a role in electron transfer in the metabolism of OHR.
Keywords: PceC; RdhC; flavin mononucleotide (FMN); flavin transferase; flavin-trafficking proteins (Ftp); flavoproteins; organohalide respiration; protein reconstitution.
Figures






References
-
- Adrian L., Löffler F. (2016). Organohalide-Respiring Bacteria. Berlin: Springer-Verlag.
-
- Bertsova Y. V., Fadeeva M. S., Kostyrko V. A., Serebryakova M. V., Baykov A. A., Bogachev A. V. (2013). Alternative pyrimidine biosynthesis protein ApbE is a flavin transferase catalyzing covalent attachment of FMN to a threonine residue in bacterial flavoproteins. J. Biol. Chem. 288 14276–14286. 10.1074/jbc.M113.455402 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

