Predicting response to checkpoint inhibitors in melanoma beyond PD-L1 and mutational burden
- PMID: 29743104
- PMCID: PMC5944039
- DOI: 10.1186/s40425-018-0344-8
Predicting response to checkpoint inhibitors in melanoma beyond PD-L1 and mutational burden
Abstract
Background: Immune checkpoint inhibitors (ICIs) have changed the clinical management of melanoma. However, not all patients respond, and current biomarkers including PD-L1 and mutational burden show incomplete predictive performance. The clinical validity and utility of complex biomarkers have not been studied in melanoma.
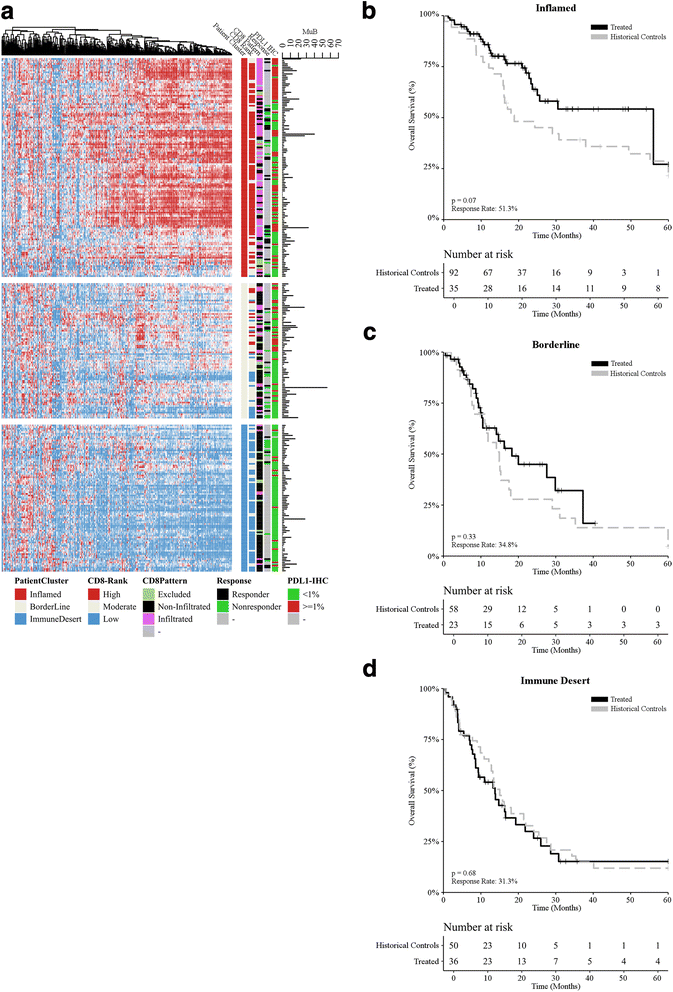
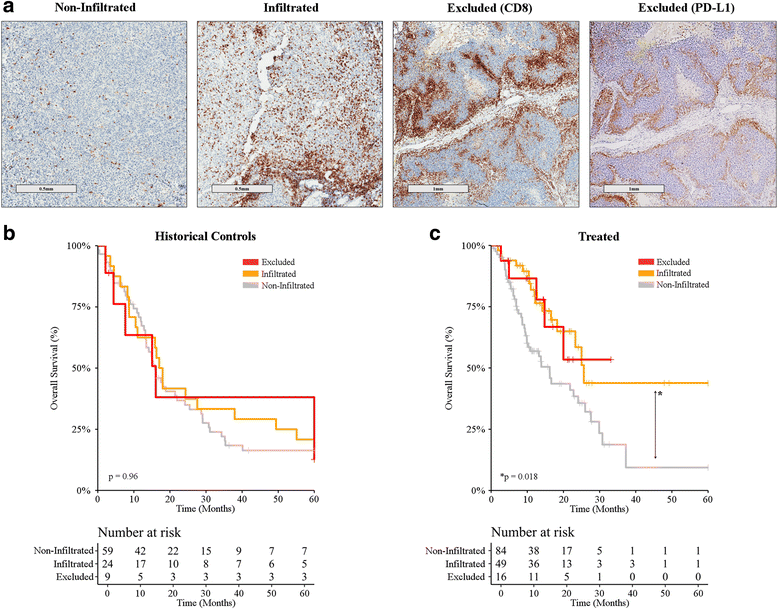
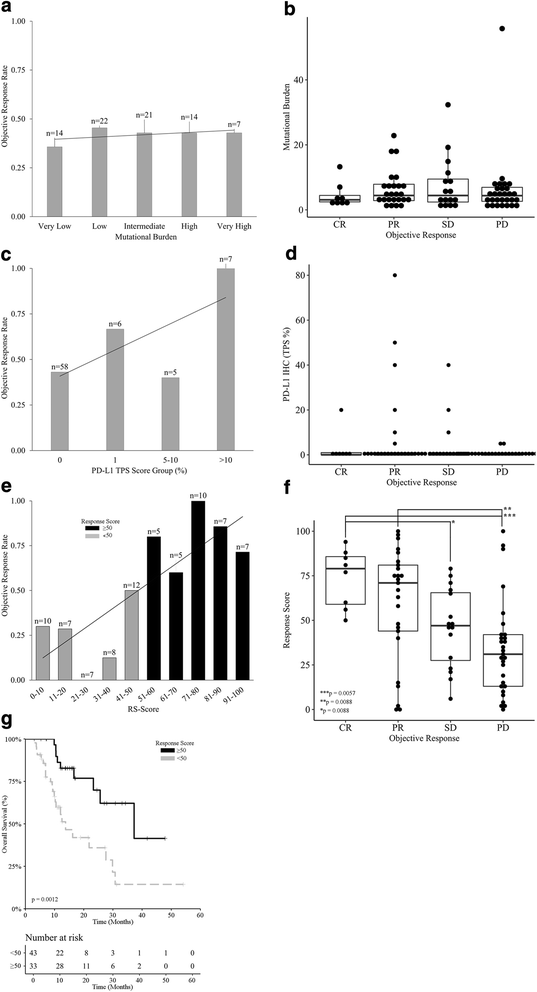
Methods: Cutaneous metastatic melanoma patients at eight institutions were evaluated for PD-L1 expression, CD8+ T-cell infiltration pattern, mutational burden, and 394 immune transcript expression. PD-L1 IHC and mutational burden were assessed for association with overall survival (OS) in 94 patients treated prior to ICI approval by the FDA (historical-controls), and in 137 patients treated with ICIs. Unsupervised analysis revealed distinct immune-clusters with separate response rates. This comprehensive immune profiling data were then integrated to generate a continuous Response Score (RS) based upon response criteria (RECIST v.1.1). RS was developed using a single institution training cohort (n = 48) and subsequently tested in a separate eight institution validation cohort (n = 29) to mimic a real-world clinical scenario.
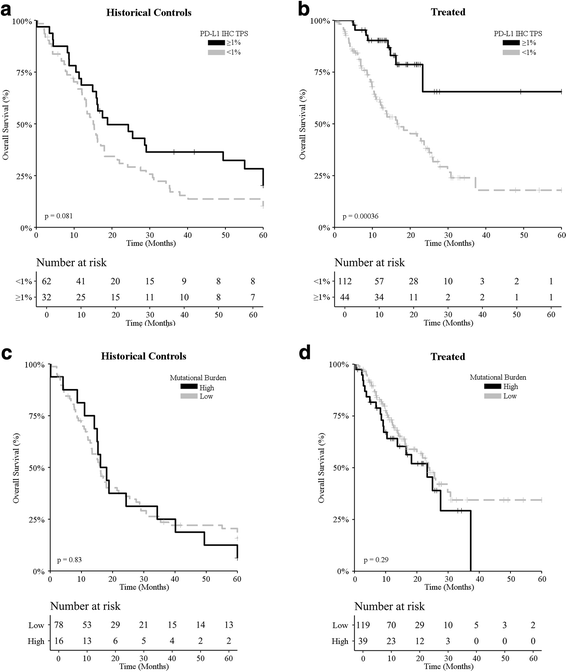
Results: PD-L1 positivity ≥1% correlated with response and OS in ICI-treated patients, but demonstrated limited predictive performance. High mutational burden was associated with response in ICI-treated patients, but not with OS. Comprehensive immune profiling using RS demonstrated higher sensitivity (72.2%) compared to PD-L1 IHC (34.25%) and tumor mutational burden (32.5%), but with similar specificity.
Conclusions: In this study, the response score derived from comprehensive immune profiling in a limited melanoma cohort showed improved predictive performance as compared to PD-L1 IHC and tumor mutational burden.
Keywords: Algorithmic analysis; Borderline; Immune Desert; Inflamed; Ipilimumab; Nivolumab; Pembrolizumab.
Conflict of interest statement
Ethics approval and consent to participate
Existing deidentified specimens and associated clinical data was submitted to OmniSeq from Roswell Park Comprehensive Cancer Center (Buffalo, NY), Fox Chase Cancer Center (Philadelphia, PA), Dartmouth-Hitchcock Medical Center (Lebanon, NH), Northwest Oncology (Munster, IN), Hospital Universitario Virgen Macarena (Sevilla, Spain), Columbia University (New York, NY), and Miami Cancer Institute (Miami, FL) with institutional review board approval from each individual institution. OmniSeq’s analysis utilized deidentified data that was considered non-human subjects research under IRB-approved protocol (BDR #073166) at Roswell Park Comprehensive Cancer Center (Buffalo, NY).
Competing interests
Disclosures: MZ, PG, KD, KS, KGM, LJT, NS, DK, LDLCM, IA, YS, MB, MVC, and ZD have no conflicts of interest to disclose. SP, JMC, MKN, STG, DD, APS, BB, JA, VG, MQ, YW, FLL, MG, and CM are all employees of OmniSeq, Inc. (Buffalo, NY) and hold restricted stock in OmniSeq, Inc. AO, WB, IP, CM, STG, JMC, and MSE are employees of Roswell Park Comprehensive Cancer Center (Buffalo, NY). Roswell Park Comprehensive Cancer Center is the majority shareholder of OmniSeq, Inc. MSE, RD, and LG provide remunerated consulting to OmniSeq, Inc. ME and SMA are employees of Laboratory Corporation of America Holdings (Burlington, NC). Laboratory Corporation of America Holdings, (Burlington, NC) is a shareholder in OmniSeq, Inc.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
