Human DHEA sulfation requires direct interaction between PAPS synthase 2 and DHEA sulfotransferase SULT2A1
- PMID: 29743239
- PMCID: PMC6016456
- DOI: 10.1074/jbc.RA118.002248
Human DHEA sulfation requires direct interaction between PAPS synthase 2 and DHEA sulfotransferase SULT2A1
Abstract
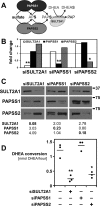
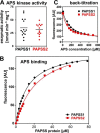
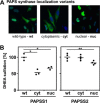
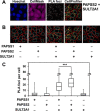
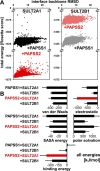
The high-energy sulfate donor 3'-phosphoadenosine-5'-phosphosulfate (PAPS), generated by human PAPS synthase isoforms PAPSS1 and PAPSS2, is required for all human sulfation pathways. Sulfotransferase SULT2A1 uses PAPS for sulfation of the androgen precursor dehydroepiandrosterone (DHEA), thereby reducing downstream activation of DHEA to active androgens. Human PAPSS2 mutations manifest with undetectable DHEA sulfate, androgen excess, and metabolic disease, suggesting that ubiquitous PAPSS1 cannot compensate for deficient PAPSS2 in supporting DHEA sulfation. In knockdown studies in human adrenocortical NCI-H295R1 cells, we found that PAPSS2, but not PAPSS1, is required for efficient DHEA sulfation. Specific APS kinase activity, the rate-limiting step in PAPS biosynthesis, did not differ between PAPSS1 and PAPSS2. Co-expression of cytoplasmic SULT2A1 with a cytoplasmic PAPSS2 variant supported DHEA sulfation more efficiently than co-expression with nuclear PAPSS2 or nuclear/cytosolic PAPSS1. Proximity ligation assays revealed protein-protein interactions between SULT2A1 and PAPSS2 and, to a lesser extent, PAPSS1. Molecular docking studies showed a putative binding site for SULT2A1 within the PAPSS2 APS kinase domain. Energy-dependent scoring of docking solutions identified the interaction as specific for the PAPSS2 and SULT2A1 isoforms. These findings elucidate the mechanistic basis for the selective requirement for PAPSS2 in human DHEA sulfation.
Keywords: DHEAS; PAPS synthase; dehydroepiandrosterone; enzyme kinetics; molecular docking; protein–protein interaction; steroid hormone; steroid sulfation pathway; sulfotransferase.
© 2018 Mueller et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

