Mitochondrial Complex I activity signals antioxidant response through ERK5
- PMID: 29743487
- PMCID: PMC5943249
- DOI: 10.1038/s41598-018-23884-4
Mitochondrial Complex I activity signals antioxidant response through ERK5
Abstract
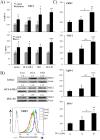
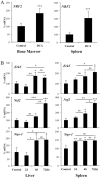
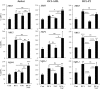
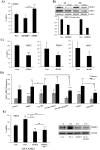
Oxidative phosphorylation (OXPHOS) generates ROS as a byproduct of mitochondrial complex I activity. ROS-detoxifying enzymes are made available through the activation of their antioxidant response elements (ARE) in their gene promoters. NRF2 binds to AREs and induces this anti-oxidant response. We show that cells from multiple origins performing OXPHOS induced NRF2 expression and its transcriptional activity. The NRF2 promoter contains MEF2 binding sites and the MAPK ERK5 induced MEF2-dependent NRF2 expression. Blocking OXPHOS in a mouse model decreased Erk5 and Nrf2 expression. Furthermore, fibroblasts derived from patients with mitochondrial disorders also showed low expression of ERK5 and NRF2 mRNAs. Notably, in cells lacking functional mitochondrial complex I activity OXPHOS did not induce ERK5 expression and failed to generate this anti-oxidant response. Complex I activity induces ERK5 expression through fumarate accumulation. Eukaryotic cells have evolved a genetic program to prevent oxidative stress directly linked to OXPHOS and not requiring ROS.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Human Leukemic Cells performing Oxidative Phosphorylation (OXPHOS) Generate an Antioxidant Response Independently of Reactive Oxygen species (ROS) Production.EBioMedicine. 2015 Nov 26;3:43-53. doi: 10.1016/j.ebiom.2015.11.045. eCollection 2016 Jan. EBioMedicine. 2015. PMID: 26870816 Free PMC article.
-
Laminar flow activation of ERK5 protein in vascular endothelium leads to atheroprotective effect via NF-E2-related factor 2 (Nrf2) activation.J Biol Chem. 2012 Nov 23;287(48):40722-31. doi: 10.1074/jbc.M112.381509. Epub 2012 Oct 5. J Biol Chem. 2012. PMID: 23043106 Free PMC article.
-
Fluvastatin inhibits AGE-induced cell proliferation and migration via an ERK5-dependent Nrf2 pathway in vascular smooth muscle cells.PLoS One. 2017 May 22;12(5):e0178278. doi: 10.1371/journal.pone.0178278. eCollection 2017. PLoS One. 2017. PMID: 28542559 Free PMC article.
-
The PDK1 Inhibitor Dichloroacetate Controls Cholesterol Homeostasis Through the ERK5/MEF2 Pathway.Sci Rep. 2017 Sep 6;7(1):10654. doi: 10.1038/s41598-017-10339-5. Sci Rep. 2017. PMID: 28878225 Free PMC article.
-
PERK regulates Nrf2/ARE antioxidant pathway against dibutyl phthalate-induced mitochondrial damage and apoptosis dependent of reactive oxygen species in mouse spermatocyte-derived cells.Toxicol Lett. 2019 Jun 15;308:24-33. doi: 10.1016/j.toxlet.2019.03.007. Epub 2019 Mar 23. Toxicol Lett. 2019. PMID: 30910607
Cited by
-
NRF2: A crucial regulator for mitochondrial metabolic shift and prostate cancer progression.Front Physiol. 2022 Sep 23;13:989793. doi: 10.3389/fphys.2022.989793. eCollection 2022. Front Physiol. 2022. PMID: 36213236 Free PMC article. Review.
-
PCK1 Downregulation Promotes TXNRD1 Expression and Hepatoma Cell Growth via the Nrf2/Keap1 Pathway.Front Oncol. 2018 Dec 17;8:611. doi: 10.3389/fonc.2018.00611. eCollection 2018. Front Oncol. 2018. PMID: 30619751 Free PMC article.
-
Regulation of Nrf2 by Mitochondrial Reactive Oxygen Species in Physiology and Pathology.Biomolecules. 2020 Feb 17;10(2):320. doi: 10.3390/biom10020320. Biomolecules. 2020. PMID: 32079324 Free PMC article. Review.
-
Hypoglycemia and hyperinsulinemia induced by phenolic uremic toxins in CKD and DKD patients.Sci Rep. 2025 Feb 17;15(1):5762. doi: 10.1038/s41598-025-87501-x. Sci Rep. 2025. PMID: 39962199 Free PMC article.
-
Preventive Action of Beta-Carotene against the Indoxyl Sulfate-Induced Renal Dysfunction in Male Adult Zebrafish via Regulations of Mitochondrial Inflammatory and β-Carotene Oxygenase-2 Actions.Biomedicines. 2023 Sep 27;11(10):2654. doi: 10.3390/biomedicines11102654. Biomedicines. 2023. PMID: 37893028 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

