Reprogramming, oscillations and transdifferentiation in epigenetic landscapes
- PMID: 29743499
- PMCID: PMC5943272
- DOI: 10.1038/s41598-018-25556-9
Reprogramming, oscillations and transdifferentiation in epigenetic landscapes
Abstract
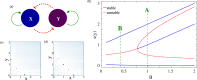
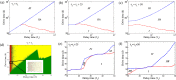
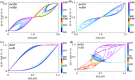
Waddington's epigenetic landscape provides a phenomenological understanding of the cell differentiation pathways from the pluripotent to mature lineage-committed cell lines. In light of recent successes in the reverse programming process there has been significant interest in quantifying the underlying landscape picture through the mathematics of gene regulatory networks. We investigate the role of time delays arising from multi-step chemical reactions and epigenetic rearrangement on the cell differentiation landscape for a realistic two-gene regulatory network, consisting of self-promoting and mutually inhibiting genes. Our work provides the first theoretical basis of the transdifferentiation process in the presence of delays, where one differentiated cell type can transition to another directly without passing through the undifferentiated state. Additionally, the interplay of time-delayed feedback and a time dependent chemical drive leads to long-lived oscillatory states in appropriate parameter regimes. This work emphasizes the important role played by time-delayed feedback loops in gene regulatory circuits and provides a framework for the characterization of epigenetic landscapes.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

