Haplotype analysis of APOE intragenic SNPs
- PMID: 29745836
- PMCID: PMC5998902
- DOI: 10.1186/s12868-018-0413-4
Haplotype analysis of APOE intragenic SNPs
Abstract
Background: APOE ε4 allele is most common genetic risk factor for Alzheimer's disease (AD) and cognitive decline. However, it remains poorly understood why only some carriers of APOE ε4 develop AD and how ethnic variabilities in APOE locus contribute to AD risk. Here, to address the role of APOE haplotypes, we reassessed the diversity of APOE locus in major ethnic groups and in Alzheimer's Disease Neuroimaging Initiative (ADNI) dataset on patients with AD, and subjects with mild cognitive impairment (MCI), and control non-demented individuals.
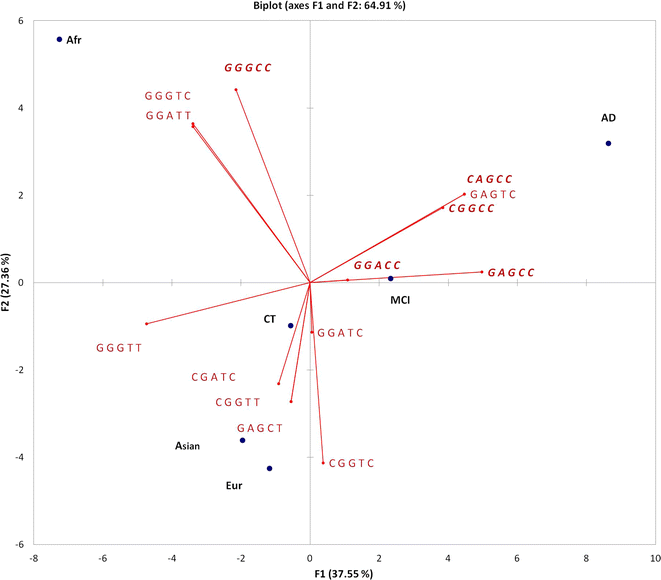
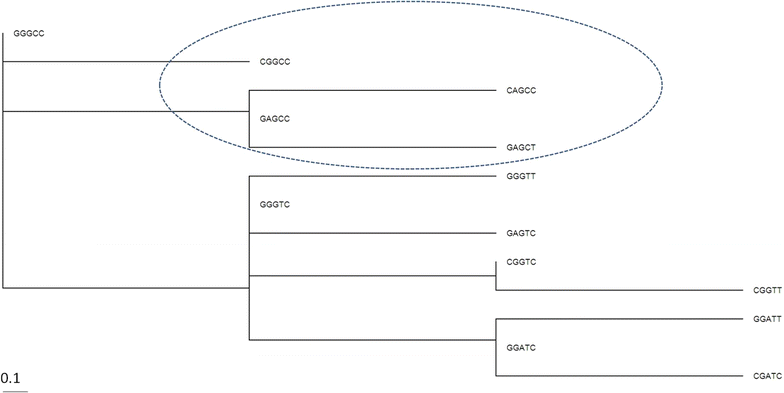
Results: We performed APOE gene haplotype analysis for a short block of five SNPs across the gene using the ADNI whole genome sequencing dataset. The compilation of ADNI data with 1000 Genomes identified the APOE ε4 linked haplotypes, which appeared to be distant for the Asian, African and European populations. The common European ε4-bearing haplotype is associated with AD but not with MCI, and the Africans lack this haplotype. Haplotypic inference revealed alleles that may confer protection against AD. By assessing the DNA methylation profile of the APOE haplotypes, we found that the AD-associated haplotype features elevated APOE CpG content, implying that this locus can also be regulated by genetic-epigenetic interactions.
Conclusions: We showed that SNP frequency profiles within APOE locus are highly skewed to population-specific haplotypes, suggesting that the ancestral background within different sites at APOE gene may shape the disease phenotype. We propose that our results can be utilized for more specific risk assessment based on population descent of the individuals and on higher specificity of five site haplotypes associated with AD.
Keywords: ADNI dataset; APOE; Alzheimer’s disease; DNA methylation; GWAS; Haplotype analysis; PCA; SNPs.
Figures




References
-
- Takei N, Miyashita A, Tsukie T, Arai H, Asada T, Imagawa M, Shoji M, Higuchi S, Urakami K, Kimura H, Kakita A, Takahashi H, Tsuji S, Kanazawa I, Ihara Y, Odani S, Kuwano R. Japanese genetic study consortium for Alzheimer Disease. Genetic association study on in and around the APOE in late-onset Alzheimer disease in Japanese. Genomics. 2009;93(5):441–448. doi: 10.1016/j.ygeno.2009.01.003. - DOI - PubMed
-
- Anderson ED, Wahoske M, Huber M, Norton D, Li Z, Koscik RL, Umucu E, Johnson SC, Jones J, Asthana S, Gleason CE. Alzheimer’s disease neuroimaging initiative. Cognitive variability-a marker for incident MCI and AD: an analysis for the Alzheimer’s Disease neuroimaging initiative. Alzheimers Dement (Amst) 2016;4:47–55. - PMC - PubMed
-
- Luo X, Qiu T, Jia Y, Huang P, Xu X, Yu X, Shen Z, Jiaerken Y, Guan X, Zhou J, Zhang M. ADNI. Intrinsic functional connectivity alterations in cognitively intact elderly APOE ε4 carriers measured by eigenvector centrality mapping are related to cognition and CSF biomarkers: a preliminary study. Brain Imaging Behav. 2017;11(5):1290–1301. doi: 10.1007/s11682-016-9600-z. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

