Multi-target drug repositioning by bipartite block-wise sparse multi-task learning
- PMID: 29745839
- PMCID: PMC5998894
- DOI: 10.1186/s12918-018-0569-7
Multi-target drug repositioning by bipartite block-wise sparse multi-task learning
Abstract
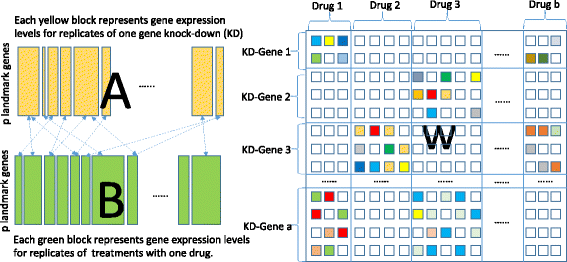
Background: Finding potential drug targets is a crucial step in drug discovery and development. Recently, resources such as the Library of Integrated Network-Based Cellular Signatures (LINCS) L1000 database provide gene expression profiles induced by various chemical and genetic perturbations and thereby make it possible to analyze the relationship between compounds and gene targets at a genome-wide scale. Current approaches for comparing the expression profiles are based on pairwise connectivity mapping analysis. However, this method makes the simple assumption that the effect of a drug treatment is similar to knocking down its single target gene. Since many compounds can bind multiple targets, the pairwise mapping ignores the combined effects of multiple targets, and therefore fails to detect many potential targets of the compounds.
Results: We propose an algorithm to find sets of gene knock-downs that induce gene expression changes similar to a drug treatment. Assuming that the effects of gene knock-downs are additive, we propose a novel bipartite block-wise sparse multi-task learning model with super-graph structure (BBSS-MTL) for multi-target drug repositioning that overcomes the restrictive assumptions of connectivity mapping analysis.
Conclusions: The proposed method BBSS-MTL is more accurate for predicting potential drug targets than the simple pairwise connectivity mapping analysis on five datasets generated from different cancer cell lines.
Availability: The code can be obtained at http://gr.xjtu.edu.cn/web/liminli/codes .
Keywords: Drug repositioning; L1000; Multi-task learning.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

