Fe-S Clusters and MutY Base Excision Repair Glycosylases: Purification, Kinetics, and DNA Affinity Measurements
- PMID: 29746241
- PMCID: PMC6267926
- DOI: 10.1016/bs.mie.2017.11.035
Fe-S Clusters and MutY Base Excision Repair Glycosylases: Purification, Kinetics, and DNA Affinity Measurements
Abstract
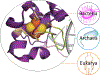
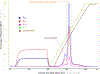
A growing number of iron-sulfur (Fe-S) cluster cofactors have been identified in DNA repair proteins. MutY and its homologs are base excision repair (BER) glycosylases that prevent mutations associated with the common oxidation product of guanine (G), 8-oxo-7,8-dihydroguanine (OG) by catalyzing adenine (A) base excision from inappropriately formed OG:A mispairs. The finding of an [4Fe-4S]2+ cluster cofactor in MutY, Endonuclease III, and structurally similar BER enzymes was surprising and initially thought to represent an example of a purely structural role for the cofactor. However, in the two decades subsequent to the initial discovery, purification and in vitro analysis of bacterial MutYs and mammalian homologs, such as human MUTYH and mouse Mutyh, have demonstrated that proper Fe-S cluster coordination is required for OG:A substrate recognition and adenine excision. In addition, the Fe-S cluster in MutY has been shown to be capable of redox chemistry in the presence of DNA. The work in our laboratory aimed at addressing the importance of the MutY Fe-S cluster has involved a battery of approaches, with the overarching hypothesis that understanding the role(s) of the Fe-S cluster is intimately associated with understanding the biological and chemical properties of MutY and its unique damaged DNA substrate as a whole. In this chapter, we focus on methods of enzyme expression and purification, detailed enzyme kinetics, and DNA affinity assays. The methods described herein have not only been leveraged to provide insight into the roles of the MutY Fe-S cluster but have also been provided crucial information needed to delineate the impact of inherited variants of the human homolog MUTYH associated with a colorectal cancer syndrome known as MUTYH-associated polyposis or MAP. Notably, many MAP-associated variants have been found adjacent to the Fe-S cluster further underscoring the intimate relationship between the cofactor, MUTYH-mediated DNA repair, and disease.
Keywords: DNA damage; DNA repair; DNA–protein interactions; Enzyme kinetics; Fe–S cluster; Protein purification.
© 2018 Elsevier Inc. All rights reserved.
Figures



References
-
- Al-Tassan N, Chmiel NH, Maynard J, Fleming N, Livingston AL, Williams GT, Hodges AK, Davies DR, David SS, Sampson JR, and Cheadle JR (2002). Inherited variants of MYH associated with somatic G : C -> T : A mutations in colorectal tumors. Nat Genet 30, 227–232. - PubMed
-
- Bai H, Jones S, Guan X, Wilson TM, Sampson JR, Cheadle JP, and Lu A-L (2005). Functional characterization of two human MutY homolog (hMYH) missense mutations (R227W and V232F) that lie within the putative hMSH6 binding domain and are associated with hMYH polyposis. Nucleic Acids Res 33, 597–604. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

