Nuclear IHC enumeration: A digital phantom to evaluate the performance of automated algorithms in digital pathology
- PMID: 29746503
- PMCID: PMC5944932
- DOI: 10.1371/journal.pone.0196547
Nuclear IHC enumeration: A digital phantom to evaluate the performance of automated algorithms in digital pathology
Abstract
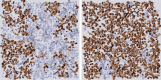
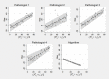
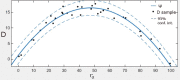
Automatic and accurate detection of positive and negative nuclei from images of immunostained tissue biopsies is critical to the success of digital pathology. The evaluation of most nuclei detection algorithms relies on manually generated ground truth prepared by pathologists, which is unfortunately time-consuming and suffers from inter-pathologist variability. In this work, we developed a digital immunohistochemistry (IHC) phantom that can be used for evaluating computer algorithms for enumeration of IHC positive cells. Our phantom development consists of two main steps, 1) extraction of the individual as well as nuclei clumps of both positive and negative nuclei from real WSI images, and 2) systematic placement of the extracted nuclei clumps on an image canvas. The resulting images are visually similar to the original tissue images. We created a set of 42 images with different concentrations of positive and negative nuclei. These images were evaluated by four board certified pathologists in the task of estimating the ratio of positive to total number of nuclei. The resulting concordance correlation coefficients (CCC) between the pathologist and the true ratio range from 0.86 to 0.95 (point estimates). The same ratio was also computed by an automated computer algorithm, which yielded a CCC value of 0.99. Reading the phantom data with known ground truth, the human readers show substantial variability and lower average performance than the computer algorithm in terms of CCC. This shows the limitation of using a human reader panel to establish a reference standard for the evaluation of computer algorithms, thereby highlighting the usefulness of the phantom developed in this work. Using our phantom images, we further developed a function that can approximate the true ratio from the area of the positive and negative nuclei, hence avoiding the need to detect individual nuclei. The predicted ratios of 10 held-out images using the function (trained on 32 images) are within ±2.68% of the true ratio. Moreover, we also report the evaluation of a computerized image analysis method on the synthetic tissue dataset.
Conflict of interest statement
Figures



References
-
- Zaha DC. Significance of immunohistochemistry in breast cancer. World journal of clinical oncology. 2014;5(3):382 doi: 10.5306/wjco.v5.i3.382 - DOI - PMC - PubMed
-
- Niazi MKK, Downs-Kelly E, Gurcan MN, editors. Hot spot detection for breast cancer in Ki-67 stained slides: image dependent filtering approach. SPIE Medical Imaging; 2014 2014: International Society for Optics and Photonics.
-
- Di Cataldo S, Ficarra E, Acquaviva A, Macii E. Automated segmentation of tissue images for computerized IHC analysis. Computer methods and programs in biomedicine. 2010;100(1):1–15. doi: 10.1016/j.cmpb.2010.02.002 - DOI - PubMed
-
- Taylor C, Levenson RM. Quantification of immunohistochemistry—issues concerning methods, utility and semiquantitative assessment II. Histopathology. 2006;49(4):411–24. doi: 10.1111/j.1365-2559.2006.02513.x - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

