Characterization of a nontypeable Haemophilus influenzae thermonuclease
- PMID: 29746527
- PMCID: PMC5944969
- DOI: 10.1371/journal.pone.0197010
Characterization of a nontypeable Haemophilus influenzae thermonuclease
Abstract
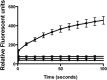
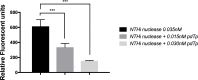
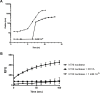
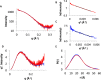
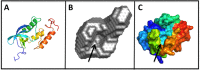
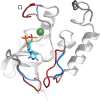
Nontypeable Haemophilus influenzae (NTHi) has been shown to form biofilms, comprised of extracellular DNA (eDNA), in the middle ear and bronchus during clinical infections. Studies in our laboratory have shown that NTHi possesses a homolog of Staphylococcus aureus thermonuclease (staphylococcal thermonuclease), NTHi nuclease (NTHi Nuc, HI_1296). This enzyme had similar size, heat stability, and divalent cation requirements to those of the staphylococcal homolog as determined by light scattering and circular dichroism spectroscopy. Small angle X-ray scattering (SAXS) analysis suggested an overall shape and substrate-binding site comparable to those of staphylococcal nuclease. However, NTHi Nuc was approximately 25-fold more active in fluorescence resonance energy transfer (FRET) activity assay than staphylococcal thermonuclease. Homology modeling implicates shorter NTHi Nuc loops near the active site for this enhanced activity.
Conflict of interest statement
Figures


References
-
- Murphy TF, Apicella MA. Nontypable Haemophilus influenzae: a review of clinical aspects, surface antigens and the human response to infection. Rev Infect Dis. 1987;9:1–15. - PubMed
-
- Chin CL, Manzel LJ, Lehman EE, Humlicek AL, Shi L, Starner TD, et al. Haemophilus influenzae from patients with chronic obstructive pulmonary disease exacerbation induce more inflammation than colonizers. Am J Respir Crit Care Med. 2005;172(1):85–91. Epub 2005/04/05. doi: 10.1164/rccm.200412-1687OC ; PubMed Central PMCID: PMC2718449. - DOI - PMC - PubMed
-
- Post JC. Direct evidence of bacterial biofilms in otitis media. Laryngoscope. 2001;111(12):2083–94. doi: 10.1097/00005537-200112000-00001 . - DOI - PubMed
-
- Starner TD, Zhang N, Kim G, Apicella MA, McCray PB Jr. Haemophilus influenzae forms biofilms on airway epithelia: implications in cystic fibrosis. Am J Respir Crit Care Med. 2006;174(2):213–20. Epub 2006/05/06. doi: 10.1164/rccm.200509-1459OC ; PubMed Central PMCID: PMC2662906. - DOI - PMC - PubMed
-
- Stoodley P, Sauer K, Davies DG, Costerton JW. Biofilms as complex differentiated communities. Annu Rev Microbiol. 2002;56:187–209. doi: 10.1146/annurev.micro.56.012302.160705 . - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

