Adaptation of S. cerevisiae to Fermented Food Environments Reveals Remarkable Genome Plasticity and the Footprints of Domestication
- PMID: 29746697
- PMCID: PMC5995190
- DOI: 10.1093/molbev/msy066
Adaptation of S. cerevisiae to Fermented Food Environments Reveals Remarkable Genome Plasticity and the Footprints of Domestication
Abstract
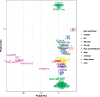
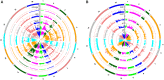
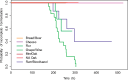
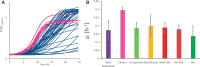
The budding yeast Saccharomyces cerevisiae can be found in the wild and is also frequently associated with human activities. Despite recent insights into the phylogeny of this species, much is still unknown about how evolutionary processes related to anthropogenic niches have shaped the genomes and phenotypes of S. cerevisiae. To address this question, we performed population-level sequencing of 82 S. cerevisiae strains from wine, flor, rum, dairy products, bakeries, and the natural environment (oak trees). These genomic data enabled us to delineate specific genetic groups corresponding to the different ecological niches and revealed high genome content variation across the groups. Most of these strains, compared with the reference genome, possessed additional genetic elements acquired by introgression or horizontal transfer, several of which were population-specific. In addition, several genomic regions in each population showed evidence of nonneutral evolution, as shown by high differentiation, or of selective sweeps including genes with key functions in these environments (e.g., amino acid transport for wine yeast). Linking genetics to lifestyle differences and metabolite traits has enabled us to elucidate the genetic basis of several niche-specific population traits, such as growth on galactose for cheese strains. These data indicate that yeast has been subjected to various divergent selective pressures depending on its niche, requiring the development of customized genomes for better survival in these environments. These striking genome dynamics associated with local adaptation and domestication reveal the remarkable plasticity of the S. cerevisiae genome, revealing this species to be an amazing complex of specialized populations.
Figures


References
-
- Alachiotis N, Stamatakis A, Pavlidis P.. 2012. OmegaPlus: a scalable tool for rapid detection of selective sweeps in whole-genome datasets. Bioinformatics 2817:2274–2275. - PubMed
-
- Almeida P, Barbosa R, Bensasson D, Gonçalves P, Sampaio JP.. 2017. Adaptive divergence in wine yeasts and their wild relatives suggests a prominent role for introgressions and rapid evolution at noncoding sites. Mol Ecol. 267:2167–2182. - PubMed
-
- Almeida P, Barbosa R, Zalar P, Imanishi Y, Shimizu K, Turchetti B, Legras J-L, Serra M, Dequin S, Couloux A et al. , . 2015. A population genomics insight into the Mediterranean origins of wine yeast domestication. Mol Ecol. 2421:5412–5427. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

