Multiproteomic and Transcriptomic Analysis of Oncogenic β-Catenin Molecular Networks
- PMID: 29747501
- PMCID: PMC7385754
- DOI: 10.1021/acs.jproteome.8b00180
Multiproteomic and Transcriptomic Analysis of Oncogenic β-Catenin Molecular Networks
Abstract
The dysregulation of Wnt signaling is a frequent occurrence in many different cancers. Oncogenic mutations of CTNNB1/β-catenin, the key nuclear effector of canonical Wnt signaling, lead to the accumulation and stabilization of β-catenin protein with diverse effects in cancer cells. Although the transcriptional response to Wnt/β-catenin signaling activation has been widely studied, an integrated understanding of the effects of oncogenic β-catenin on molecular networks is lacking. We used affinity-purification mass spectrometry (AP-MS), label-free liquid chromatography-tandem mass spectrometry, and RNA-Seq to compare protein-protein interactions, protein expression, and gene expression in colorectal cancer cells expressing mutant (oncogenic) or wild-type β-catenin. We generate an integrated molecular network and use it to identify novel protein modules that are associated with mutant or wild-type β-catenin. We identify a DNA methyltransferase I associated subnetwork that is enriched in cells with mutant β-catenin and a subnetwork enriched in wild-type cells associated with the CDKN2A tumor suppressor, linking these processes to the transformation of colorectal cancer cells through oncogenic β-catenin signaling. In summary, multiomics analysis of a defined colorectal cancer cell model provides a significantly more comprehensive identification of functional molecular networks associated with oncogenic β-catenin signaling.
Keywords: DNA methyltransferase I; Wnt signaling; multiomics integration; oncogenic mutations; protein−protein interaction network; β-catenin.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
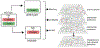




References
-
- Clevers H Wnt/Beta-Catenin Signaling in Development and Disease. Cell 2006, 127 (3), 469–480. - PubMed
-
- Morin PJ; Sparks AB; Korinek V; Barker N; Clevers H; Vogelstein B; Kinzler KW Activation of Beta-Catenin-Tcf Signaling in Colon Cancer by Mutations in Beta-Catenin or APC. Science 1997, 275 (5307), 1787–1790. - PubMed
-
- Angers S; Moon RT Proximal Events in Wnt Signal Transduction. Nat. Rev. Mol. Cell Biol. 2009, 10, 468. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

