MicroRNA signature of central nervous system-infiltrating dendritic cells in an animal model of multiple sclerosis
- PMID: 29749614
- PMCID: PMC6099169
- DOI: 10.1111/imm.12934
MicroRNA signature of central nervous system-infiltrating dendritic cells in an animal model of multiple sclerosis
Abstract
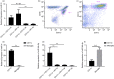
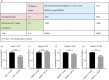
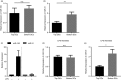
Innate immune cells are integral to the pathogenesis of several diseases of the central nervous system (CNS), including multiple sclerosis (MS). Dendritic cells (DCs) are potent CD11c+ antigen-presenting cells that are critical regulators of adaptive immune responses, particularly in autoimmune diseases such as MS. The regulation of DC function in both the periphery and CNS compartment has not been fully elucidated. One limitation to studying the role of CD11c+ DCs in the CNS is that microglia can upregulate CD11c during inflammation, making it challenging to distinguish bone marrow-derived DCs (BMDCs) from microglia. Selective expression of microRNAs (miRNAs) has been shown to distinguish populations of innate cells and regulate their function within the CNS during neuro-inflammation. Using the experimental autoimmune encephalomyelitis (EAE) murine model of MS, we characterized the expression of miRNAs in CD11c+ cells using a non-biased murine array. Several miRNAs, including miR-31, were enriched in CD11c+ cells within the CNS during EAE, but not LysM+ microglia. Moreover, to distinguish CD11c+ DCs from microglia that upregulate CD11c, we generated bone marrow chimeras and found that miR-31 expression was specific to BMDCs. Interestingly, miR-31-binding sites were enriched in mRNAs downregulated in BMDCs that migrated into the CNS, and a subset was confirmed to be regulated by miR-31. Finally, miR-31 was elevated in DCs migrating through an in vitro blood-brain barrier. Our findings suggest miRNAs, including miR-31, may regulate entry of DCs into the CNS during EAE, and could potentially represent therapeutic targets for CNS autoimmune diseases such as MS.
Keywords: dendritic cells; microRNAs; multiple sclerosis; neuroinflammation.
© 2018 John Wiley & Sons Ltd.
Figures

References
-
- Frohman EM, Racke MK, Raine CS. Multiple sclerosis–the plaque and its pathogenesis. N Engl J Med 2006; 354:942–55. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

