Population-specific genetic modification of Huntington's disease in Venezuela
- PMID: 29750799
- PMCID: PMC5965898
- DOI: 10.1371/journal.pgen.1007274
Population-specific genetic modification of Huntington's disease in Venezuela
Abstract
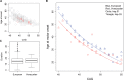
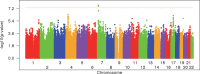
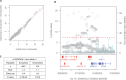
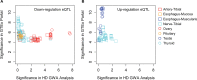
Modifiers of Mendelian disorders can provide insights into disease mechanisms and guide therapeutic strategies. A recent genome-wide association (GWA) study discovered genetic modifiers of Huntington's disease (HD) onset in Europeans. Here, we performed whole genome sequencing and GWA analysis of a Venezuelan HD cluster whose families were crucial for the original mapping of the HD gene defect. The Venezuelan HD subjects develop motor symptoms earlier than their European counterparts, implying the potential for population-specific modifiers. The main Venezuelan HD family inherits HTT haplotype hap.03, which differs subtly at the sequence level from European HD hap.03, suggesting a different ancestral origin but not explaining the earlier age at onset in these Venezuelans. GWA analysis of the Venezuelan HD cluster suggests both population-specific and population-shared genetic modifiers. Genome-wide significant signals at 7p21.2-21.1 and suggestive association signals at 4p14 and 17q21.2 are evident only in Venezuelan HD, but genome-wide significant association signals at the established European chromosome 15 modifier locus are improved when Venezuelan HD data are included in the meta-analysis. Venezuelan-specific association signals on chromosome 7 center on SOSTDC1, which encodes a bone morphogenetic protein antagonist. The corresponding SNPs are associated with reduced expression of SOSTDC1 in non-Venezuelan tissue samples, suggesting that interaction of reduced SOSTDC1 expression with a population-specific genetic or environmental factor may be responsible for modification of HD onset in Venezuela. Detection of population-specific modification in Venezuelan HD supports the value of distinct disease populations in revealing novel aspects of a disease and population-relevant therapeutic strategies.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Bates GP, Dorsey R, Gusella JF, Hayden MR, Kay C, et al. (2015) Huntington disease. Nature Reviews Disease Primers: 15005 doi: 10.1038/nrdp.2015.5 - DOI - PubMed
-
- Huntington G (1872) On chorea. Med Surg Rep 26: 320–321.
-
- The Huntington's Disease Collaborative Research Group (1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 72: 971–983. - PubMed
-
- Gilliam TC, Bucan M, MacDonald ME, Zimmer M, Haines JL, et al. (1987) A DNA segment encoding two genes very tightly linked to Huntington's disease. Science 238: 950–952. - PubMed
-
- Gilliam TC, Tanzi RE, Haines JL, Bonner TI, Faryniarz AG, et al. (1987) Localization of the Huntington's disease gene to a small segment of chromosome 4 flanked by D4S10 and the telomere. Cell 50: 565–571. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

