Resistance to pentamidine is mediated by AdeAB, regulated by AdeRS, and influenced by growth conditions in Acinetobacter baumannii ATCC 17978
- PMID: 29750823
- PMCID: PMC5947904
- DOI: 10.1371/journal.pone.0197412
Resistance to pentamidine is mediated by AdeAB, regulated by AdeRS, and influenced by growth conditions in Acinetobacter baumannii ATCC 17978
Abstract
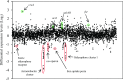
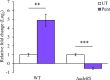
In recent years, effective treatment of infections caused by Acinetobacter baumannii has become challenging due to the ability of the bacterium to acquire or up-regulate antimicrobial resistance determinants. Two component signal transduction systems are known to regulate expression of virulence factors including multidrug efflux pumps. Here, we investigated the role of the AdeRS two component signal transduction system in regulating the AdeAB efflux system, determined whether AdeA and/or AdeB can individually confer antimicrobial resistance, and explored the interplay between pentamidine resistance and growth conditions in A. baumannii ATCC 17978. Results identified that deletion of adeRS affected resistance towards chlorhexidine and 4',6-diamidino-2-phenylindole dihydrochloride, two previously defined AdeABC substrates, and also identified an 8-fold decrease in resistance to pentamidine. Examination of ΔadeA, ΔadeB and ΔadeAB cells augmented results seen for ΔadeRS and identified a set of dicationic AdeAB substrates. RNA-sequencing of ΔadeRS revealed transcription of 290 genes were ≥2-fold altered compared to the wildtype. Pentamidine shock significantly increased adeA expression in the wildtype, but decreased it in ΔadeRS, implying that AdeRS activates adeAB transcription in ATCC 17978. Investigation under multiple growth conditions, including the use of Biolog phenotypic microarrays, revealed resistance to pentamidine in ATCC 17978 and mutants could be altered by bioavailability of iron or utilization of different carbon sources. In conclusion, the results of this study provide evidence that AdeAB in ATCC 17978 can confer intrinsic resistance to a subset of dicationic compounds and in particular, resistance to pentamidine can be significantly altered depending on the growth conditions.
Conflict of interest statement
Figures


References
-
- Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21(3):538–82. doi: 10.1128/CMR.00058-07 - DOI - PMC - PubMed
-
- Fournier PE, Vallenet D, Barbe V, Audic S, Ogata H, Poirel L, et al. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2006;2(1):e7 doi: 10.1371/journal.pgen.0020007 - DOI - PMC - PubMed
-
- Adams MD, Chan ER, Molyneaux ND, Bonomo RA. Genomewide analysis of divergence of antibiotic resistance determinants in closely related isolates of Acinetobacter baumannii. Antimicrob Agents Chemother. 2010;54(9):3569–77. doi: 10.1128/AAC.00057-10 - DOI - PMC - PubMed
-
- Blackwell GA, Hamidian M, Hall RM. IncM plasmid R1215 is the source of chromosomally located regions containing multiple antibiotic resistance genes in the globally disseminated Acinetobacter baumannii GC1 and GC2 clones. mSphere. 2016;1(3):e00117–16. doi: 10.1128/mSphere.00117-16 - DOI - PMC - PubMed
-
- Hamidian M, Kenyon JJ, Holt KE, Pickard D, Hall RM. A conjugative plasmid carrying the carbapenem resistance gene blaOXA-23 in AbaR4 in an extensively resistant GC1 Acinetobacter baumannii isolate. J Antimicrob Chemother. 2014;69(10):2625–8. doi: 10.1093/jac/dku188 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

