Whole Exome Sequencing Identifies New Host Genomic Susceptibility Factors in Empyema Caused by Streptococcus pneumoniae in Children: A Pilot Study
- PMID: 29751582
- PMCID: PMC5977180
- DOI: 10.3390/genes9050240
Whole Exome Sequencing Identifies New Host Genomic Susceptibility Factors in Empyema Caused by Streptococcus pneumoniae in Children: A Pilot Study
Abstract
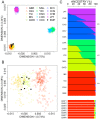
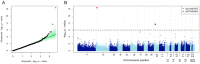
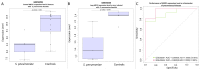
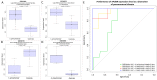
Pneumonia is the leading cause of death amongst infectious diseases. Streptococcus pneumoniae is responsible for about 25% of pneumonia cases worldwide, and it is a major cause of childhood mortality. We carried out a whole exome sequencing (WES) study in eight patients with complicated cases of pneumococcal pneumonia (empyema). An initial assessment of statistical association of WES variation with pneumonia was carried out using data from the 1000 Genomes Project (1000G) for the Iberian Peninsula (IBS) as reference controls. Pseudo-replication statistical analyses were carried out using different European control groups. Association tests pointed to single nucleotide polymorphism (SNP) rs201967957 (gene MEIS1; chromosome 2; p-valueIBS = 3.71 × 10-13) and rs576099063 (gene TSPAN15; chromosome 10; p-valueIBS = 2.36 × 10-8) as the best candidate variants associated to pneumococcal pneumonia. A burden gene test of pathogenicity signaled four genes, namely, OR9G9, MUC6, MUC3A and APOB, which carry significantly increased pathogenic variation when compared to controls. By analyzing various transcriptomic data repositories, we found strong supportive evidence for the role of MEIS1, TSPAN15 and APOBR (encoding the receptor of the APOB protein) in pneumonia in mouse and human models. Furthermore, the association of the olfactory receptor gene OR9G9 has recently been related to some viral infectious diseases, while the role of mucin genes (MUC6 and MUC3A), encoding mucin glycoproteins, are well-known factors related to chronic obstructive airway disease. WES emerges as a promising technique to disentangle the genetic basis of host genome susceptibility to infectious respiratory diseases.
Keywords: Streptococcus pneumoniae; infectious disease; next generation sequencing; parallel sequencing; pediatrics; transcriptome; whole exome sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- O’Brien K.L., Wolfson L.J., Watt J.P., Henkle E., Deloria-Knoll M., McCall N., Lee E., Mulholland K., Levine O.S., Cherian T., et al. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: Global estimates. Lancet. 2009;374:893–902. doi: 10.1016/S0140-6736(09)61204-6. - DOI - PubMed
-
- Liu L., Oza S., Hogan D., Chu Y., Perin J., Zhu J., Lawn J.E., Cousens S., Mathers C., Black R.E. Global, regional, and national causes of under-5 mortality in 2000–15: An updated systematic analysis with implications for the sustainable development goals. Lancet. 2016;388:3027–3035. doi: 10.1016/S0140-6736(16)31593-8. - DOI - PMC - PubMed
-
- Dion C.F., Ashurst J.V. Pneumonia, Streptococcus Pneumoniae. StatPearls Publising; Treasure Island, FL, USA: 2018.
-
- Martinón-Torres F., Dosil-Gallardo S., Pérez-del-Molino-Bernal M.L., Sánchez F.P., Tarrago D., Álvez F., Díaz S.P., Martinón-Torres N., Martinón-Sánchez J.M. Pleural antigen assay in the diagnosis of pediatric pneumococcal empyema. J. Crit. Care. 2012;27:321. doi: 10.1016/j.jcrc.2011.05.004. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

