The Sucrose Synthase Gene Family in Chinese Pear (Pyrus bretschneideri Rehd.): Structure, Expression, and Evolution
- PMID: 29751599
- PMCID: PMC6100409
- DOI: 10.3390/molecules23051144
The Sucrose Synthase Gene Family in Chinese Pear (Pyrus bretschneideri Rehd.): Structure, Expression, and Evolution
Abstract
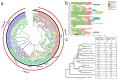
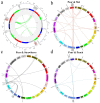
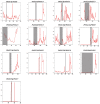
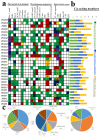
Sucrose synthase (SS) is a key enzyme involved in sucrose metabolism that is critical in plant growth and development, and particularly quality of the fruit. Sucrose synthase gene families have been identified and characterized in plants various plants such as tobacco, grape, rice, and Arabidopsis. However, there is still lack of detailed information about sucrose synthase gene in pear. In the present study, we performed a systematic analysis of the pear (Pyrus bretschneideri Rehd.) genome and reported 30 sucrose synthase genes. Subsequently, gene structure, phylogenetic relationship, chromosomal localization, gene duplications, promoter regions, collinearity, RNA-Seq data and qRT-PCR were conducted on these sucrose synthase genes. The transcript analysis revealed that 10 PbSSs genes (30%) were especially expressed in pear fruit development. Additionally, qRT-PCR analysis verified the RNA-seq data and shown that PbSS30, PbSS24, and PbSS15 have a potential role in the pear fruit development stages. This study provides important insights into the evolution of sucrose synthase gene family in pear and will provide assistance for further investigation of sucrose synthase genes functions in the process of fruit development, fruit quality and resistance to environmental stresses.
Keywords: RNA-seq data analysis; characteristics; expression pattern; phylogenetic analysis; sucrose synthase.
Conflict of interest statement
The authors declare they have no competing interests.
Figures


References
-
- Hirose T., Scofield G.N., Terao T. An expression analysis profile for the entire sucrose synthase gene family in rice. Plant Sci. 2008;174:534–543. doi: 10.1016/j.plantsci.2008.02.009. - DOI
-
- Islam M.Z., Hu X.-M., Jin L.-F., Liu Y.-Z., Peng S.-A. Genome-wide identification and expression profile analysis of citrus sucrose synthase genes: Investigation of possible roles in the regulation of sugar accumulation. PLoS ONE. 2014;9:e113623. doi: 10.1371/journal.pone.0113623. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

