Natronobiforma cellulositropha gen. nov., sp. nov., a novel haloalkaliphilic member of the family Natrialbaceae (class Halobacteria) from hypersaline alkaline lakes
- PMID: 29752017
- PMCID: PMC6052348
- DOI: 10.1016/j.syapm.2018.04.002
Natronobiforma cellulositropha gen. nov., sp. nov., a novel haloalkaliphilic member of the family Natrialbaceae (class Halobacteria) from hypersaline alkaline lakes
Erratum in
-
Corrigendum to "Natronobiforma cellulositropha gen. nov., sp. nov., a novel haloalkaliphilic member of the family Natrialbaceae (class Halobacteria) from hypersaline alkaline lakes" [Syst. Appl. Microbiol. 41 (2018) 355-362].Syst Appl Microbiol. 2019 Mar;42(2):261-262. doi: 10.1016/j.syapm.2018.11.007. Epub 2018 Dec 11. Syst Appl Microbiol. 2019. PMID: 30551955 Free PMC article. No abstract available.
Abstract
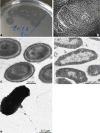
Six strains of extremely halophilic and alkaliphilic euryarchaea were enriched and isolated in pure culture from surface brines and sediments of hypersaline alkaline lakes in various geographical locations with various forms of insoluble cellulose as growth substrate. The cells are mostly flat motile rods with a thin monolayer cell wall while growing on cellobiose. In contrast, the cells growing with cellulose are mostly nonmotile cocci covered with a thick external EPS layer. The isolates, designated AArcel, are obligate aerobic heterotrophs with a narrow substrate spectrum. All strains can use insoluble celluloses, cellobiose, a few soluble glucans and xylan as their carbon and energy source. They are extreme halophiles, growing within the range from 2.5 to 4.8M total Na+ (optimum at 4M) and obligate alkaliphiles, with the pH range for growth from 7.5 to 9.9 (optimum at 8.5-9). The core archaeal lipids of strain AArcel5T were dominated by C20-C20 dialkyl glycerol ether (DGE) (i.e. archaeol) and C20-C25 DGE in nearly equal proportion. The 16S rRNA gene analysis indicated that all six isolates belong to a single genomic species mostly related to the genera Saliphagus-Natribaculum-Halovarius. Taking together a substantial phenotypic difference of the new isolates from the closest relatives and the phylogenetic distance, it is concluded that the AArcel group represents a novel genus-level branch within the family Natrialbaceae for which the name Natronobiforma cellulositropha gen. nov., sp. nov. is proposed with AArcel5T as the type strain (JCM 31939T=UNIQEM U972T).
Keywords: Cellulotrophic; Haloalkaliphilic; Hypersaline; Natronoarchaea; Soda lakes.
Copyright © 2018 The Authors. Published by Elsevier GmbH.. All rights reserved.
Figures


Similar articles
-
Natrarchaeobius chitinivorans gen. nov., sp. nov., and Natrarchaeobius halalkaliphilus sp. nov., alkaliphilic, chitin-utilizing haloarchaea from hypersaline alkaline lakes.Syst Appl Microbiol. 2019 May;42(3):309-318. doi: 10.1016/j.syapm.2019.01.001. Epub 2019 Jan 8. Syst Appl Microbiol. 2019. PMID: 30638904 Free PMC article.
-
Halococcoides cellulosivorans gen. nov., sp. nov., an extremely halophilic cellulose-utilizing haloarchaeon from hypersaline lakes.Int J Syst Evol Microbiol. 2019 May;69(5):1327-1335. doi: 10.1099/ijsem.0.003312. Epub 2019 Feb 25. Int J Syst Evol Microbiol. 2019. PMID: 30801242
-
Natronolimnobius sulfurireducens sp. nov. and Halalkaliarchaeum desulfuricum gen. nov., sp. nov., the first sulfur-respiring alkaliphilic haloarchaea from hypersaline alkaline lakes.Int J Syst Evol Microbiol. 2019 Sep;69(9):2662-2673. doi: 10.1099/ijsem.0.003506. Int J Syst Evol Microbiol. 2019. PMID: 31166158
-
Natronotalea proteinilytica gen. nov., sp. nov. and Longimonas haloalkaliphila sp. nov., extremely haloalkaliphilic members of the phylum Rhodothermaeota from hypersaline alkaline lakes.Int J Syst Evol Microbiol. 2017 Oct;67(10):4161-4167. doi: 10.1099/ijsem.0.002272. Epub 2017 Sep 18. Int J Syst Evol Microbiol. 2017. PMID: 28920839
-
Natronocalculus amylovorans gen. nov., sp. nov., and Natranaeroarchaeum aerophilus sp. nov., dominant culturable amylolytic natronoarchaea from hypersaline soda lakes in southwestern Siberia.Syst Appl Microbiol. 2022 Jul;45(4):126336. doi: 10.1016/j.syapm.2022.126336. Epub 2022 May 20. Syst Appl Microbiol. 2022. PMID: 35644061
Cited by
-
Natrarchaeobius chitinivorans gen. nov., sp. nov., and Natrarchaeobius halalkaliphilus sp. nov., alkaliphilic, chitin-utilizing haloarchaea from hypersaline alkaline lakes.Syst Appl Microbiol. 2019 May;42(3):309-318. doi: 10.1016/j.syapm.2019.01.001. Epub 2019 Jan 8. Syst Appl Microbiol. 2019. PMID: 30638904 Free PMC article.
-
Symbiosis between nanohaloarchaeon and haloarchaeon is based on utilization of different polysaccharides.Proc Natl Acad Sci U S A. 2020 Aug 18;117(33):20223-20234. doi: 10.1073/pnas.2007232117. Epub 2020 Aug 5. Proc Natl Acad Sci U S A. 2020. PMID: 32759215 Free PMC article.
-
Selective enrichment on a wide polysaccharide spectrum allowed isolation of novel metabolic and taxonomic groups of haloarchaea from hypersaline lakes.Front Microbiol. 2022 Nov 23;13:1059347. doi: 10.3389/fmicb.2022.1059347. eCollection 2022. Front Microbiol. 2022. PMID: 36504804 Free PMC article.
-
Haloarchaea as Promising Chassis to Green Chemistry.Microorganisms. 2024 Aug 22;12(8):1738. doi: 10.3390/microorganisms12081738. Microorganisms. 2024. PMID: 39203580 Free PMC article. Review.
-
Halophilic archaea and their potential to generate renewable fuels and chemicals.Biotechnol Bioeng. 2021 Mar;118(3):1066-1090. doi: 10.1002/bit.27639. Epub 2020 Dec 16. Biotechnol Bioeng. 2021. PMID: 33241850 Free PMC article. Review.
References
-
- Andrei A.S., Banciu H.L., Oren A. Living with salt: metabolic and phylogenetic diversity of archaea inhabiting saline ecosystems. FEMS Microbiol. Lett. 2012;330:1–9. - PubMed
-
- Amoozegar M.A., Siroosi M., Atashgahi S., Smidt H., Ventosa A. Systematics of haloarchaea and biotechnological potential of their hydrolytic enzymes. Microbiology. 2017;163:623–645. - PubMed
-
- Bhatnagar T., Boutaiba S., Hacene H., Cayol J.-L., Fardeau M.-L., Ollivier B., Baratti J.C. Lipolytic activity from Halobacteria: screening and hydrolase production. FEMS Microbiol. Lett. 2005;248:133–140. - PubMed
-
- Castillo A.M., Gutiérrez M.C., Kamekura M., Ma Y., Cowan D.A., Jones B.E., Grant W.D., Ventosa A. Halovivax asiaticus gen. nov., sp. nov., a novel extremely halophilic archaeon isolated from Inner Mongolia, China. Int. J. Syst. Evol. Microbiol. 2006;56:765–770. - PubMed
-
- Enache M., Kamekura M. Hydrolytic enzymes of halophilic microorganisms and their economic values. Rom. J. Biochem. 2010;47:47–59.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

